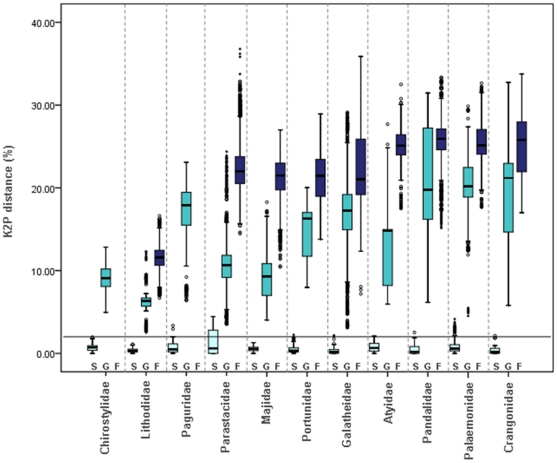
Figure 3. Boxplot distribution of 11 selected families of the Decapoda order intraspecies (S), intragenus (G), and intrafamily (F) COI K2P distances (%).
The plot summarises median (central bar), position of the upper and lower quartiles (called Q1 and Q3, central box), extremes of the data (dots) and very extreme points of the distribution that can be considered as outliers (stars). Points are considered as outliers when they exceed Q3+1.5(Q3-Q1) for the lower part, where (Q3-Q1) is the inter quartile range. The number of sequences, species, and genera per family are given in Table 3. Mean K2P distance (%) ± SE within taxa are: Chirostylidae S = 0.701±0.028 and G = 8.999±.039; Lithodidae S = 0.416±0.021, G = 6.376±.137 and F = 11.392±0.063; Paguridae S = 0.686±.045 and G = 17.173±0.084; Parastacidae S = 1.375±0.131, G = 11.017±0.078 and F = 22.681±0.064; Majidae S = 0.547±0.028, G = 9.643±0.214 and F = 21.084±0.061; Portunidae S = 0.453±0.024, G = 14.826±0.311 and F = 28.929±0.047; Galatheidae S = 0.285±0.017, G = 16.839±0.04 and F = 22.355±0.033; Atyidae S = 0.758±0.041, G = 13.475±0.352 and F = 25.218±0.056; Pandalidae S = 0.49±0.042, G = 20.924±0.213 and F = 25.617±0.07; Palaemonidae S = 0.812±0.055, G = 20.157±0.108 and F = 25.398±0.048; Crangonidae S = 0.344, G = 19.991±0.514 and F = 25.241±0.103.