Abstract
Quinacrine, a drug with antimalarial and anticancer activities that inhibits NF-κB and activates p53, has progressed into phase II clinical trials in cancer. To further elucidate its mechanism of action and identify pathways of drug resistance, we used an unbiased method for validation-based insertional mutagenesis to isolate a quinacrine-resistant cell line in which an inserted CMV promoter drives overexpression of the FER tyrosine kinase (FER). Overexpression of FER from a cDNA confers quinacrine resistance to several different types of cancer cell lines. We show that quinacrine kills cancer cells primarily by inhibiting the activation of NF-κB and that increased activation of NF-κB through FER overexpression mediates resistance. EGF activates NF-κB and stimulates phosphorylation of FER, EGF receptor (EGFR), and ERK p42/p44, and decreased expression of FER or inhibition of ERK phosphorylation inhibits the EGF-induced activation of NF-κB. FER binds to EGFR, and overexpression of FER in cells untreated with EGF increases this association, leading to increased phosphorylation of EGFR and ERK. We conclude that FER is on a pathway connecting EGFR to NF-κB activation and that this function is responsible for FER-dependent resistance to quinacrine.
Quinacrine has been used in humans for many years to treat malaria, autoimmune disorders, and other conditions (1–3). Quinacrine simultaneously activates p53 and inhibits activated NF-κB, making it a very promising anticancer drug (4, 5). To further elucidate its mechanism of action as an anticancer agent and identify pathways of resistance, we used validation-based insertional mutagenesis (VBIM) to generate mutant cells that resist killing by quinacrine. In VBIM, the strong CMV promoter is inserted into many different loci in the genomes of a population of mammalian cells, causing increased expression of downstream genes (6). The inserted promoter can be excised to prove that the altered phenotype has been caused by the insertion. As a powerful tool for genetic study, VBIM has recently been applied to different selections, with productive outcomes (6, 7).
NF-κB is activated by numerous external stimuli and has a major role in inducing inflammation. The canonical NF-κB complex of p65 and p50 subunits is sequestered in the cytoplasm through its association with the inhibitory subunit IκB (8). After stimulation with cytokines such as TNFα or IL-1, IκB kinase (IKK) is activated, which leads to the phosphorylation of IκB, targeting it for proteosome-mediated degradation and liberating NF-κB (8). Activated EGF also drives NF-κB activation, but the details of this pathway are not yet well-understood (9–11). The binding of EGF to its receptor (EGFR), a receptor tyrosine kinase, leads to EGFR dimerization and autophosphorylation, and then, it leads to activation of downstream signaling pathways (12). FER, a tyrosine kinase that is activated by cell-surface receptors such as EGFR, platelet-derived growth factor receptor (PDGFR), and FcγR after ligand engagement (13–15), has an N-terminal FER-CIP4 homology (FCH) domain, three coiled coils, a central SH2 domain, and a carboxyl-terminal kinase domain (16). Activated FER associates with and activates cellular proteins containing SH2 domains (17–20). We have now found that FER is on a pathway through which EGF activates NF-κB and that overexpression of FER activates NF-κB, thus conferring resistance to the NF-κB inhibitor quinacrine.
Results
Identification of FER in a Quinacrine-Resistant Clone.
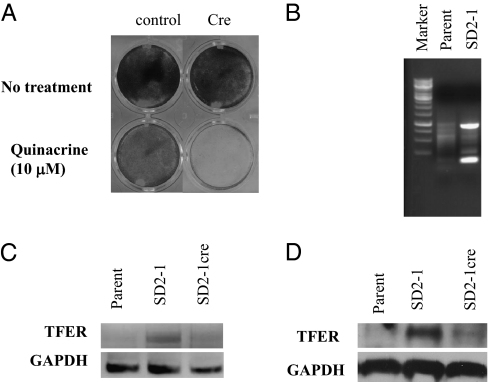
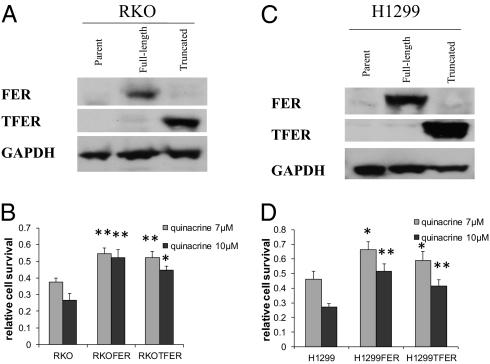
Eighteen different pools of human colon cancer RKO cells were infected with three different VBIM viruses (6) using a total of 1 million cells. After propagation, each pool was replated and treated with 10 μM quinacrine for 48 h. Twenty quinacrine-resistant colonies were observed 2 wk later in seven of the pools. The VBIM vectors contain LoxP sites, allowing excision of the promoter in candidate mutant clones. We infected each clone with a vector expressing Cre recombinase (6) followed by treatment with 10 μM quinacrine for 48 h. The quinacrine-resistant phenotype was reversed in mutant SD2-1 (Fig. 1A), showing that the inserted CMV promoter caused the resistance in this case. We used two-step inverse PCR (iPCR) to identify the sequences flanking the insertion site in mutant SD2-1 (Materials and Methods). Two iPCR products were cloned and sequenced (Fig. 1B). The sequences flanking one product are located on human chromosome 5q21 in intron 10 of the FER gene. The sequences flanking the other PCR product did not match any sequence in the database. The full coding sequence of FER translates to 822 aa, and the insertion leads to the expression of a truncated protein (TFER) with 356 aa, which still includes the complete SH2 and kinase domains. An analysis of mRNA revealed that TFER is expressed in mutant SD2-1 cells (Fig. 1C), and a Western analysis showed the expression of TFER protein in these cells (Fig. 1D). We expressed FER or TFER in unmutagenized colon cancer RKO cells or lung cancer H1299 cells (Fig. 2 A and C) and tested their sensitivity to quinacrine. Overexpression of either protein confers resistance to both cell types (Fig. 2 B and D). The results in H1299 cells, which lack expression of p53 (1), show that the ability of FER to mediate quinacrine resistance does not depend on p53.
Fig. 1.
Identification of FER in a quinacrine-resistant clone. (A) Clone SD2-1 cells were infected with control vector (pBabepuro) or a vector encoding Cre recombinase. The cells were plated into 24-well plates at 35,000 cells/well. The next day, the cells were treated with quinacrine (10 μM) and stained with methylene blue 48 h later. (B) Genomic DNA from parental RKO or mutant SD2-1 cells was isolated. After digestion and self-ligation (Materials and Methods), the products were used as templates in a PCR to identify the genomic sequences flanking the insertion site. (C) Truncated FER (TFER) mRNA levels were analyzed by RT-PCR using 5′ VBIM-specific and 3′ gene-specific primers that allow amplification of TFER mRNA in parental RKO, SD2-1, or SD2-1cre cells. GAPDH was used as a loading control. (D) TFER protein expression in parental RKO, SD2-1, or SD2-1cre cells was analyzed by the Western method using an FER-specific antibody with GAPDH as a loading control.
Fig. 2.
Effects of FER overexpression on cell proliferation and sensitivity to quinacrine. (A) RKO or (C) H1299 cells were infected with an empty vector or a vector encoding FER or TFER, and their expression was examined by the Western method with GAPDH as a loading control. (B) RKO or (D) H1299 cells were treated with the indicated concentrations of quinacrine for 24 h, and then grown in drug-free medium for another 24 h. Cell survival was measured by the Cell Titer Blue method. The data shown represent means ± SD from three experiments. *P < 0.05; **P < 0.005.
Up-Regulation of NF-κB Activity in Cells Overexpressing FER.
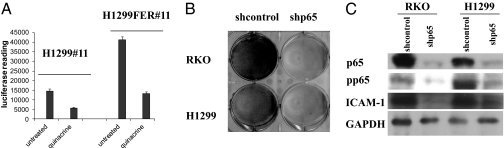
NF-κB activation is a common mechanism through which tumors develop resistance to chemotherapy (21), and one of the primary activities of quinacrine is to inhibit the constitutive activation of NF-κB (1, 22). Therefore, we investigated the role of FER in regulating NF-κB activation in tumor cells. H1299 cells with an integrated NF-κB–dependent luciferase reporter (H1299#11 cells) were transduced with a vector expressing FER to create H1299FER#11 cells. In these cells, the overexpression of FER increased basal NF-κB–dependent luciferase activity (Fig. 3A). When parental cells or cells overexpressing FER were treated with quinacrine, this activity was decreased (Fig. 3A).
Fig. 3.
Overexpression of FER activates NF-κB, and NF-κB is essential for cancer cell survival. (A) Overexpression of FER activates NF-κB. H1299#11 or H1299FER#11 cells were treated with quinacrine (10 μM) for 24 h, and NF-κB–dependent luciferase activity was measured 24 h later. The cell lysates were normalized for protein concentration. The results of triplicate luciferase assays are shown as means ± SD. (B) NF-κB is essential for cancer cell survival. RKO and H1299 cells were infected with a vector encoding a control shRNA or shRNAs against the p65 subunit of NF-κB. The cells were stained with methylene blue after infection. (C) The expression of p65, phospho-(S536)p65, and the NF-κB target gene ICAM-1 was analyzed by the Western method with GAPDH as a loading control.
NF-κB Is Essential for Cancer Cell Survival.
NF-κB is activated constitutively in many types of tumors (23), inducing the expression of NF-κB–dependent genes whose products inhibit apoptosis and promote proliferation, inflammation, and invasion (24). To show in the RKO colon cancer cells and H1299 lung cancer cells that we have studied that the constitutive activation of NF-κB is essential for survival, these cells were infected with a viral vector encoding an shRNA against the p65 subunit of NF-κB. Loss of p65 expression led to the death of both types of cells (Fig. 3B). p65 knockdown also decreased the phosphorylation of p65 on Ser536 and expression of the p65 target gene intercellular adhesion molecule 1 (ICAM-1) (25) in both cell types (Fig. 3C). Similar results were reported previously by Gurova et al. (4).
Overexpression of FER Does Not Activate the AKT Pathway.
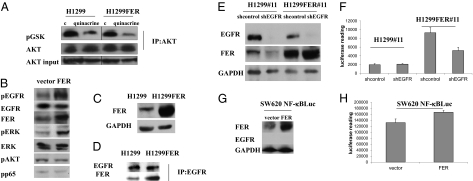
It has been reported that the overexpression of FER increases the association of insulin receptor substrate-1 with the p85 subunit of PI3K (19). Stimulation with PDGF also results in the formation of a complex between FER, PDGFR, and p85 (20). Because activation of the PI3K/AKT pathway is known to activate NF-κB (26), we tested whether FER mediates NF-κB activation through this pathway. H1299 and H1299FER cells were treated with quinacrine, and AKT activity was analyzed in an in vitro kinase assay using the AKT substrate glycogen synthase kinase (GSK). Overexpression of FER did not affect AKT activity, and quinacrine treatment decreased AKT activity to the same extent in H1299 and H1299FER cells (Fig. 4A). Also, FER overexpression did not increase the phosphorylation of AKT (Fig. 4B). These results show that FER does not activate the PI3K/AKT pathway. Therefore, some other mechanism is responsible for the activation of NF-κB in response to FER overexpression.
Fig. 4.
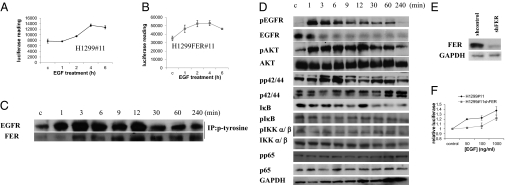
FER overexpression in the absence of EGF increases the phosphorylation of EGFR and ERK but not AKT; EGFR is important for FER-mediated activation of NF-κB. (A) FER overexpression does not activate the PI3K/AKT pathway. H1299 and H1299FER cells were treated with quinacrine (10 μM) for 24 h, cell lysates were immunoprecipitated with an antibody to AKT, and the products were analyzed in an in vitro kinase assay, with recombinant GSK as substrate, using an antibody to phosphorylated GSK. (B) FER overexpression induced the phosphorylation of EGFR and ERK. H1299 cells were transfected transiently with a control vector or a vector expressing FER. Cell lysates were analyzed by the Western method 48 h after transfection. (C and D) Interaction of FER with EGFR. (C) FER overexpression in H1299 FER cells was analyzed by the Western method with GAPDH as a loading control. (D) Extracts of H1299 and H1299FER cells were immunoprecipitated with an antibody to EGFR, and the products were analyzed by the Western method. (E and F) H1299#11 and H1299FER#11 cells were infected with vectors encoding a control shRNA or shRNAs against EGFR. (E) The levels of EGFR and FER were analyzed by the Western method with GAPDH as a loading control. (F) NF-κB–dependent luciferase activity was measured after infection. The lysates were normalized for protein concentration. The results of triplicate luciferase assays are shown as means ± SD. (G and H) SW620 cells with an integrated NF-κB–dependent luciferase reporter were infected with vectors encoding control or FER cDNAs. (G) The levels of EGFR and FER were examined by the Western method with GAPDH as a loading control. (H) NF-κB–dependent luciferase activity was measured after infection. The lysates were normalized for protein concentration. The results of triplicate luciferase assays are shown as means ± SD.
FER and EGF-Dependent Signaling.
Treatment of cells with EGF and PDGF stimulates FER phosphorylation (13, 14). Treatment with EGF can also activate NF-κB (27, 28), and several lines of evidence indicate that aberrant EGFR activation correlates with constitutively active NF-κB in cancer. In breast cancer cell lines, NF-κB activation has been shown to be downstream of EGFR-dependent signaling, especially in estrogen receptor (ER)-negative cells (9, 10). In prostate cancer cells, EGFR has been reported to regulate the constitutive activation of NF-κB (11). To determine whether FER is involved in the activation of NF-κB in EGF-treated cells, we analyzed the effect of FER on the tyrosine phosphorylation of EGFR, finding that FER overexpression greatly increases EGFR phosphorylation (Fig. 4B). Because the activation of ERK is a major downstream response to EGF and because the kinase activity of FER sustains ERK activation (29), we analyzed the phosphorylation status of ERK after FER overexpression. ERK phosphorylation was increased by FER overexpression, but the level of ERK protein did not change (Fig. 4B). Phosphorylation of the p65 subunit of NF-κB on Ser536 contributes importantly to the transactivation of NF-κB in response to cytokines (30), but overexpression of FER did not affect the phosphorylation of this residue, suggesting that FER does not activate NF-κB through this pathway (Fig. 4B). To test whether EGFR and FER bind to each other constitutively, EGFR was immunoprecipitated from extracts of untreated H1299 or H1299FER cells and analyzed by the Western method. FER was associated with EGFR in H1299 cells, and there was more association in H1299FER cells (Fig. 4 C and D). These results indicate that FER overexpression increases its association with EGFR and activates EGFR, even without EGF treatment.
We also tested whether EGFR is needed for FER to activate NF-κB. H1299#11 and H1299FER#11 cells were infected with a control shRNA or an shRNA against EGFR, and NF-κB activity was measured (Fig. 4 E and F). Knockdown of EGFR decreased NF-κB activity in cells overexpressing FER but not in control H1299 cells. In SW620 cells, a colon cancer cell line that lacks EGFR expression (31), FER overexpression activates NF-κB activity only slightly (Fig. 4 G and H). These results suggest that EGFR plays a major role in the activation of NF-κB in response to FER overexpression.
We then analyzed the kinetics of EGF-dependent activation of EGFR, FER, and NF-κB. Treatment of H1299#11 and H1299FER#11 cells with EGF led to NF-κB activation in a time-dependent manner (Fig. 5 A and B). The results shown in Fig. 5B indicate that FER overexpression greatly potentiates the ability of EGF to activate NF-κB. To monitor EGFR and FER activation, we measured the tyrosine phosphorylation of these proteins. H1299 cells were treated with EGF, cell lysates were immunoprecipitated with an antibody against phosphorylated tyrosine, and the immunocomplexes were analyzed by the Western method. Tyrosine phosphorylation of EGFR was apparent 1 min after treatment, but tyrosine phosphorylation of FER was not seen until 3 min (Fig. 5C). As expected, treatment with EGF also increased the phosphorylation of AKT and ERK p42/44 (Fig. 5D). We also examined how the canonical NF-κB pathway responds to EGF stimulation. There was no significant change in the phosphorylation of IKKα/β on Ser176/180, the phosphorylation of IκB on Ser32/36, or the phosphorylation of p65 on Ser536 (Fig. 5D). We did detect decreased expression of IκB at late times, suggesting that EGF-induced activation of NF-κB might involve eventual degradation of IκB that is not mediated through the phosphorylation of Ser32 and Ser36 (Fig. 5D). To test the role of FER in the EGF-mediated activation of NF-κB more directly, the expression of FER was inhibited by shRNA in H1299#11 cells (Fig. 5E) followed by stimulation with EGF. EGF increased NF-κB–dependent activity in a dose-dependent manner in control cells, but when FER expression was knocked down, the cells did not activate NF-κB activity as well (Fig. 5F), confirming that FER is involved in EGF-dependent NF-κB activation. Pretreatment with EGF also prevented quinacrine-mediated inhibition of NF-κB slightly (Fig. S1) but not enough to protect the cells from quinacrine-mediated cell death (Fig. S2).
Fig. 5.
Knockdown of FER inhibits EGFR activation. (A) H1299#11 or (B) H1299FER#11 cells were stimulated with EGF (500 ng/mL), and NF-κB activity was measured in a luciferase assay. The cell lysates were normalized for protein concentration. The results of triplicate luciferase assays are shown as means ± SD. (C) H1299 cells were stimulated with EGF (100 ng/mL). Cell lysates were immunoprecipitated with an antibody to phosphotyrosine and then analyzed by the Western method. (D) H1299 cells were stimulated with EGF (100 ng/mL), and cell lysates were analyzed by the Western method. (E and F) H1299#11 cells were infected with a vector encoding a control shRNA or shRNA against FER. (E) The expression levels of FER were examined by the Western method with GAPDH as a loading control. (F) The cells were treated with EGF for 6 h. After treatment, NF-κB activity was measured. The cell lysates were normalized by protein concentration. The results of triplicate luciferase assays are shown as means ± SD.
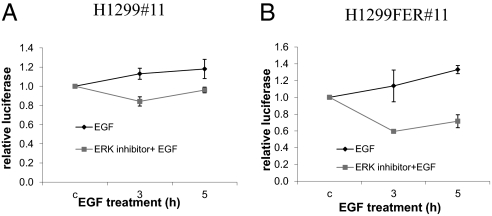
Because ERK is activated either by FER overexpression or EGF stimulation, we tested the effect of inhibiting ERK on EGF-induced NF-κB activation in H1299#11 and H1299FER #11 cells. Pretreatment with an ERK inhibitor reduced EGF-induced activation of NF-κB in both cells, but the inhibition of NF-κB is much more profound in H1299FER#11 than H1299 cells (Fig. 6 A and B).
Fig. 6.
Inhibition of ERK blocks FER- and EGF-induced activation of NF-κB. (A) H1299#11 or (B) H1299FER#11 cells were treated with PD98059 (100 μM) for 2 h and then with EGF (100 ng/mL) for the indicated times before measuring NF-κB activity. The cell lysates were normalized by protein concentration. The results of triplicate luciferase assays are shown as means ± SD.
Discussion
Development of drug resistance is a major problem in cancer treatment, and it is almost universally the case that treatment with a single drug leads to resistance (32). Resistance can occur through several different mechanisms, including amplification or mutation of a target gene and increased expression of proteins that transport the drug out of treated cells (32, 33). In the VBIM method, introduction of a CMV promoter can induce the overexpression of downstream genes, and if increased expression of the corresponding protein facilitates survival, the cells will be resistant to the drug. Previously, we used VBIM to show that the overexpression of kinesins mediates resistance to docetaxel (7), and here, we use this method to isolate a quinacrine-resistant clone with an insertion in the FER gene (Fig. 1B). We then confirm that FER overexpression confers resistance to quinacrine in naïve cells without mutagenesis (Fig. 2 B and D).
FER is expressed ubiquitously in a variety of tissues and cells (16) and is expressed at a higher level in numerous malignant cell lines than the corresponding normal cells (34–36). FER expression has been linked to prostate cancer, with higher levels in extracts from cancers compared with normal or hyperplasic tissues (34, 35). Overexpression of Drosophila FER caused malignant transformation of rodent fibroblasts (37).
Here, we show that FER overexpression activates NF-κB (Fig. 3A), helping to explain its oncogenic properties. Constitutive activation of NF-κB, apparent in most tumors, activates genes whose products are essential for cancer cell survival and proliferation, and many tumors cannot tolerate inhibition of activated NF-κB. We have also shown that knockdown of the p65 subunit of NF-κB kills different lines of cancer cells (Fig. 3 B and C), showing that NF-κB expression is necessary for their survival.
EGF plays an important role in cell proliferation and survival by activating downstream of signaling pathways involving PI3K/AKT and ERK (38). Ligand binding to EGFR leads to receptor dimerization, autophosphorylation, and activation of downstream pathways. Mutant EGFRs exhibit ligand-independent dimerization and activation of downstream signaling pathways (38). We show here that EGFR binds to FER and that this binding is increased in H1299FER cells (Fig. 4C).
We have also found that overexpression of FER increases the tyrosine phosphorylation of EGFR, the phosphorylation of ERK, and the activation of NF-κB, even in the absence of EGF (Fig. 4B). Although AKT is a downstream target of EGF-dependent signaling, we did not see AKT activation in response to FER overexpression (Fig. 4A), indicating that AKT is not involved in the FER-mediated activation of NF-κB. Consistent with this result, overexpression of HER2, an EGFR family member, also did not activate NF-κB through the AKT pathway (39).
Treatment with EGF stimulated rapid and dramatic activation of EGFR and FER, which was revealed by tyrosine phosphorylation and activation of NF-κB. Because the tyrosine phosphorylation of EGFR is apparent as soon as 1 min after treatment and the dramatic tyrosine phosphorylation of FER is seen slightly later (after 3 min) (Fig. 5B), we propose that FER may be activated by EGFR directly after binding of the SH2 domain of FER to one or more phosphotyrosine residues of EGFR after treatment with EGF. We also detected association of FER with EGFR even in the absence of treatment. FER overexpression increases its association with EGFR and activates EGFR (Fig. 4B), indicating that FER and EGFR can activate each other. We also show that inhibition of ERK activity inhibits the ability of EGF to induce the activation of NF-κB (Fig. 6A). All of these results indicate that FER is a component of an EGFR to NF-κB signaling pathway (Figs. 5D and 6 A and B). Further work is required to reveal additional components of this pathway.
Quinacrine is currently being evaluated as a therapeutic agent for several types of cancer (1, 5). Our finding that activation of EGFR induces quinacrine resistance may indicate that quinacrine may be more effective in patients with normal levels of EGFR and FER than in patients with overexpression of these proteins. It may also be advantageous to combine an ERK inhibitor with quinacrine for therapy.
Materials and Methods
Cells and Reagents.
H1299, RKO, and 293T cells were obtained from ATCC. H1299, H1299#11, and 293T cells were maintained in DMEM supplemented with 5% heat-inactivated FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin. RKO cells were maintained in RPMI medium supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin. All cultures were kept in 5% CO2 at 37 °C. Antibodies against FER, AKT, phospho-EGFR (Tyr1068), phospho-ERK (Thr202/Tyr204), ERK, phospho-AKT (Ser473), ICAM-1, phospho-IKKα/β (Ser176/180), and phospho-p65 (Ser536) were from Cell Signaling. Antibodies against p65, IκB, IKKα/β, phospho-IκB (Ser32/36), and GAPDH were from Santa Cruz Biotechnology. The antibody against EGFR was from Bethyl, and the antibody against phosphotyrosine was from BD Biosciences. The ERK inhibitor PD098059 was from EMD Chemicals.
Constructs, Virus Production, and Infection.
Human or mouse FER cDNAs were used interchangeably. They gave very similar results when compared in parallel experiments. The cDNAs were subcloned in the p-CMV-SV-40-puro lentivector (40). PLA-NF-κBLuc (41) was a gift from Peter Chumakov (Cleveland Clinic, Cleveland, OH). shRNAs against FER and p65 were from Sigma-Aldrich. To produce infectious virus, 293T cells were transfected transiently with pCMVDR8.2 and pVSV-G helper plasmids (5) as well as the plasmid of interest by using Lipofectamine Plus (Invitrogen). The virus produced was collected 24, 48, and 72 h after infection and concentrated using 40% polybrene followed by centrifugation at 2,800 × g for 20 min. To obtain stable populations, target cells were infected with virus and then selected for 2 wk.
Mapping the Inserts.
Mutant cells were cultured to 80% confluency in p100 plates. Genomic DNA was isolated from the cells using the blood and cell culture DNA midi Kit from Qiagen, digested with EcoRI and MfeI, and self-ligated with DNA ligase. The ligation products were subjected to nested PCR (6, 7).
Cell Survival Assay.
Cells were plated into 24-well plates at 30,000 cells/well. The next day, the cells were treated with quinacrine for 24 h and allowed to grow for another 24 h. Then, Cell Titer Blue (Promega) was added to the medium at a ratio of 1:5, the cells were incubated at 37 °C for 1–4 h, and the fluorescence intensity was measured.
Immunoblotting and mRNA Expression.
Cells were cultured to 80% confluency in six-well plates. After treatment, the cells were lysed with RIPA buffer (25 mM Tris·HCl, pH 7.6, 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS) containing protease inhibitor mixture (Sigma-Aldrich) and Halt phosphatase inhibitor mixture (Thermo Scientific). The lysate was centrifuged at 11,000 × g for 20 min. The concentrations of proteins in the supernatant solutions were measured by using a protein assay kit (Bio-Rad). Protein samples (20 μg) were separated in NuPAGE 4–12% electrophoresis gels (Invitrogen) and transferred to polyvinylidene difluoride membranes; immunoblot was performed according to the guidelines for each specific antibody. To determine the mRNA expression levels, total cellular RNAs were extracted by using the RNeasy Mini Kit (Qiagen). The reverse transcription (RT) reaction was performed by using the SuperScript II First-Strand Synthesis system (Invitrogen), and PCR was done with TFER-specific primers.
Luciferase Reporter Assay.
H1299#11 cells were infected with virus expressing FER or the corresponding control virus. Two days after infection, cells were replated in 12-well plates. The next day, the cells were treated with quinacrine (10 μM) overnight or EGF (100 ng/mL) for various times. After treatment, the cells were harvested in reporter lysis buffer (Promega). A 5-μL aliquot of the supernatant solution was assayed with the luciferase assay system (Promega) after brief centrifugation at 11,000 × g. The luciferase activity was normalized for protein concentration.
Immunoprecipitation.
H1299 cells were cultured to 80% confluency in 10-cm plates and then treated with EGF (100 ng/mL). Cells were lysed with immunoprecipitation (IP) buffer (20 mM Tris·HCl, pH 8, 137 mM NaCl, 10% glycerol, 1% Nonidet p-40, 2 mM EDTA) containing protease inhibitor mixture (Sigma) and Halt phosphatase inhibitor mixture (Thermo Scientific). After agitation for 30 min, the cells were centrifuged at 11,000 × g for 20 min. Equivalent amounts of supernatant solutions were mixed with an antibody against phospho-tyrosine at 4 °C overnight, with rotation. Immunocomplexes were precipitated with protein A/G agarose beads (Santa Cruz). The beads were washed four times with IP buffer, and the immunoprecipitates were analyzed by the Western method using specific antibodies.
AKT in Vitro Kinase Assay.
The AKT in vitro kinase assay kit from Cell Signaling was used according to the manufacturer's protocol.
Statistical Analysis.
Values were expressed as means ± SD. P values were based on the t test, and the significance was set at 0.05 (marked with an asterisk wherever data are statistically significant).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant CA095851 (to G.R.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105369108/-/DCSupplemental.
References
- 1.Wallace DJ. The use of quinacrine (Atabrine) in rheumatic diseases: A reexamination. Semin Arthritis Rheum. 1989;18:282–296. doi: 10.1016/0049-0172(89)90050-4. [DOI] [PubMed] [Google Scholar]
- 2.Toubi E, et al. The reduction of serum B-lymphocyte activating factor levels following quinacrine add-on therapy in systemic lupus erythematosus. Scand J Immunol. 2006;63:299–303. doi: 10.1111/j.1365-3083.2006.01737.x. [DOI] [PubMed] [Google Scholar]
- 3.Sun S, Rao NL, Venable J, Thurmond R, Karlsson L. TLR7/9 antagonists as therapeutics for immune-mediated inflammatory disorders. Inflamm Allergy Drug Targets. 2007;6:223–235. doi: 10.2174/187152807783334300. [DOI] [PubMed] [Google Scholar]
- 4.Gurova KV, et al. Small molecules that reactivate p53 in renal cell carcinoma reveal a NF-kappaB-dependent mechanism of p53 suppression in tumors. Proc Natl Acad Sci USA. 2005;102:17448–17453. doi: 10.1073/pnas.0508888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo C, et al. 9-Aminoacridine-based anticancer drugs target the PI3K/AKT/mTOR, NF-kappaB and p53 pathways. Oncogene. 2009;28:1151–1161. doi: 10.1038/onc.2008.460. [DOI] [PubMed] [Google Scholar]
- 6.Lu T, et al. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NFkappaB. Proc Natl Acad Sci USA. 2009;106:16339–16344. doi: 10.1073/pnas.0908560106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De S, Cipriano R, Jackson MW, Stark GR. Overexpression of kinesins mediates docetaxel resistance in breast cancer cells. Cancer Res. 2009;69:8035–8042. doi: 10.1158/0008-5472.CAN-09-1224. [DOI] [PubMed] [Google Scholar]
- 8.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 9.Biswas DK, Cruz AP, Gansberger E, Pardee AB. Epidermal growth factor-induced nuclear factor kappa B activation: A major pathway of cell-cycle progression in estrogen-receptor negative breast cancer cells. Proc Natl Acad Sci USA. 2000;97:8542–8547. doi: 10.1073/pnas.97.15.8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biswas DK, Iglehart JD. Linkage between EGFR family receptors and nuclear factor kappaB (NF-kappaB) signaling in breast cancer. J Cell Physiol. 2006;209:645–652. doi: 10.1002/jcp.20785. [DOI] [PubMed] [Google Scholar]
- 11.Le Page C, Koumakpayi IH, Lessard L, Mes-Masson AM, Saad F. EGFR and Her-2 regulate the constitutive activation of NF-kappaB in PC-3 prostate cancer cells. Prostate. 2005;65:130–140. doi: 10.1002/pros.20234. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto I. Epidermal growth factor receptor in relation to tumor development: EGFR-targeted anticancer therapy. FEBS J. 2010;277:309–315. doi: 10.1111/j.1742-4658.2009.07449.x. [DOI] [PubMed] [Google Scholar]
- 13.Kim L, Wong TW. Growth factor-dependent phosphorylation of the actin-binding protein cortactin is mediated by the cytoplasmic tyrosine kinase FER. J Biol Chem. 1998;273:23542–23548. doi: 10.1074/jbc.273.36.23542. [DOI] [PubMed] [Google Scholar]
- 14.Kim L, Wong TW. The cytoplasmic tyrosine kinase FER is associated with the catenin-like substrate pp120 and is activated by growth factors. Mol Cell Biol. 1995;15:4553–4561. doi: 10.1128/mcb.15.8.4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penhallow RC, Class K, Sonoda H, Bolen JB, Rowley RB. Temporal activation of nontransmembrane protein-tyrosine kinases following mast cell Fc epsilon RI engagement. J Biol Chem. 1995;270:23362–23365. doi: 10.1074/jbc.270.40.23362. [DOI] [PubMed] [Google Scholar]
- 16.Greer P. Closing in on the biological functions of Fps/Fes and fer. Nat Rev Mol Cell Biol. 2002;3:278–289. doi: 10.1038/nrm783. [DOI] [PubMed] [Google Scholar]
- 17.Taler M, et al. Fer is a downstream effector of insulin and mediates the activation of signal transducer and activator of transcription 3 in myogenic cells. Mol Endocrinol. 2003;17:1580–1592. doi: 10.1210/me.2002-0328. [DOI] [PubMed] [Google Scholar]
- 18.Zoubeidi A, et al. The Fer tyrosine kinase cooperates with interleukin-6 to activate signal transducer and activator of transcription 3 and promote human prostate cancer cell growth. Mol Cancer Res. 2009;7:142–155. doi: 10.1158/1541-7786.MCR-08-0117. [DOI] [PubMed] [Google Scholar]
- 19.Iwanishi M. Overexpression of Fer increases the association of tyrosine-phosphorylated IRS-1 with P85 phosphatidylinositol kinase via SH2 domain of Fer in transfected cells. Biochem Biophys Res Commun. 2003;311:780–785. doi: 10.1016/j.bbrc.2003.10.050. [DOI] [PubMed] [Google Scholar]
- 20.Iwanishi M, Czech MP, Cherniack AD. The protein-tyrosine kinase fer associates with signaling complexes containing insulin receptor substrate-1 and phosphatidylinositol 3-kinase. J Biol Chem. 2000;275:38995–39000. doi: 10.1074/jbc.M006665200. [DOI] [PubMed] [Google Scholar]
- 21.Luo JL, Kamata H, Karin M. IKK/NF-kappaB signaling: Balancing life and death—a new approach to cancer therapy. J Clin Invest. 2005;115:2625–2632. doi: 10.1172/JCI26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jani TS, DeVecchio J, Mazumdar T, Agyeman A, Houghton JA. Inhibition of NF-κB signaling by quinacrine is cytotoxic to human colon carcinoma cell lines and is synergistic in combination with tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) or oxaliplatin. J Biol Chem. 2010;285:19162–19172. doi: 10.1074/jbc.M109.091645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karin M, Greten FR. NF-kappaB: Linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 24.Baldwin AS., Jr. The NF-kappa B and I kappa B proteins: New discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 25.Nakanishi C, Toi M. Nuclear factor-kappaB inhibitors as sensitizers to anticancer drugs. Nat Rev Cancer. 2005;5:297–309. doi: 10.1038/nrc1588. [DOI] [PubMed] [Google Scholar]
- 26.Ozes ON, et al. NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature. 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 27.Habib AA, et al. The epidermal growth factor receptor engages receptor interacting protein and nuclear factor-kappa B (NF-kappa B)-inducing kinase to activate NF-kappa B. Identification of a novel receptor-tyrosine kinase signalosome. J Biol Chem. 2001;276:8865–8874. doi: 10.1074/jbc.M008458200. [DOI] [PubMed] [Google Scholar]
- 28.Sethi G, Ahn KS, Chaturvedi MM, Aggarwal BB. Epidermal growth factor (EGF) activates nuclear factor-kappaB through IkappaBalpha kinase-independent but EGF receptor-kinase dependent tyrosine 42 phosphorylation of IkappaBalpha. Oncogene. 2007;26:7324–7332. doi: 10.1038/sj.onc.1210544. [DOI] [PubMed] [Google Scholar]
- 29.Salem Y, et al. Fer kinase sustains the activation level of ERK1/2 and increases the production of VEGF in hypoxic cells. Cell Signal. 2005;17:341–353. doi: 10.1016/j.cellsig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W. IkappaB kinases phosphorylate NF-kappaB p65 subunit on serine 536 in the transactivation domain. J Biol Chem. 1999;274:30353–30356. doi: 10.1074/jbc.274.43.30353. [DOI] [PubMed] [Google Scholar]
- 31.Balin-Gauthier D, et al. In vivo and in vitro antitumor activity of oxaliplatin in combination with cetuximab in human colorectal tumor cell lines expressing different level of EGFR. Cancer Chemother Pharmacol. 2006;57:709–718. doi: 10.1007/s00280-005-0123-3. [DOI] [PubMed] [Google Scholar]
- 32.Luqmani YA. Mechanisms of drug resistance in cancer chemotherapy. Med Princ Pract. 2005;14(Suppl 1):35–48. doi: 10.1159/000086183. [DOI] [PubMed] [Google Scholar]
- 33.Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 34.Allard P, et al. Links between Fer tyrosine kinase expression levels and prostate cell proliferation. Mol Cell Endocrinol. 2000;159:63–77. doi: 10.1016/s0303-7207(99)00205-1. [DOI] [PubMed] [Google Scholar]
- 35.Pasder O, Salem Y, Yaffe E, Shpungin S, Nir U. FER as a novel target for cancer therapy. Drugs of the Future. 2007;32:61–70. [Google Scholar]
- 36.Li H, et al. Identification of tyrosine-phosphorylated proteins associated with metastasis and functional analysis of FER in human hepatocellular carcinoma cells. BMC Cancer. 2009;9:366–382. doi: 10.1186/1471-2407-9-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulson R, Jackson J, Immergluck K, Bishop JM. The DFer gene of Drosophila melanogaster encodes two membrane-associated proteins that can both transform vertebrate cells. Oncogene. 1997;14:641–652. doi: 10.1038/sj.onc.1200875. [DOI] [PubMed] [Google Scholar]
- 38.Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J. 2010;277:301–308. doi: 10.1111/j.1742-4658.2009.07448.x. [DOI] [PubMed] [Google Scholar]
- 39.Merkhofer EC, Cogswell P, Baldwin AS. Her2 activates NF-kappaB and induces invasion through the canonical pathway involving IKKalpha. Oncogene. 2010;29:1238–1248. doi: 10.1038/onc.2009.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheon H, Stark GR. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci USA. 2009;106:9373–9378. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gasparian AV, et al. Targeting transcription factor NFkappaB: Comparative analysis of proteasome and IKK inhibitors. Cell Cycle. 2009;8:1559–1566. doi: 10.4161/cc.8.10.8415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.