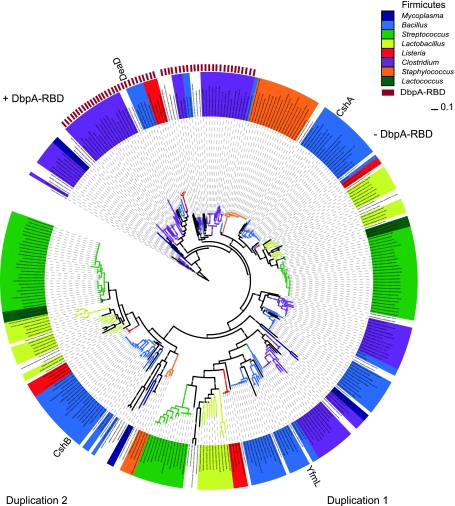
Fig. 3.
Bayesian inference of DEAD-box protein from Firmicutes. Complete alignment of 348 amino acid sequences from Firmicutes, which corresponds to the DEAD and Helicase_C domains without Gblocks filtering used in the Bayesian inference (see Fig. 1 and “Materials and methods”). DEAD-box proteins from B. subtilis are localized as reference sequences across the phylogram. The presence of the DbpA domain is shown with a red box on the perimeter. Notably it only occurs in a few genera, such as in Bacillus, Lysteria, and Clostridium. Proposed gene duplications are shown in the main clusters as duplication 1 and duplication 2. Interestingly, the Clostridium species appears to diverge earlier from other Firmicutes. The general clusters are shown in different colors and only values above 0.80 of posterior probability are shown. DbpA-RBD lacking branches next to DpbA-RBD containing branches are indicated