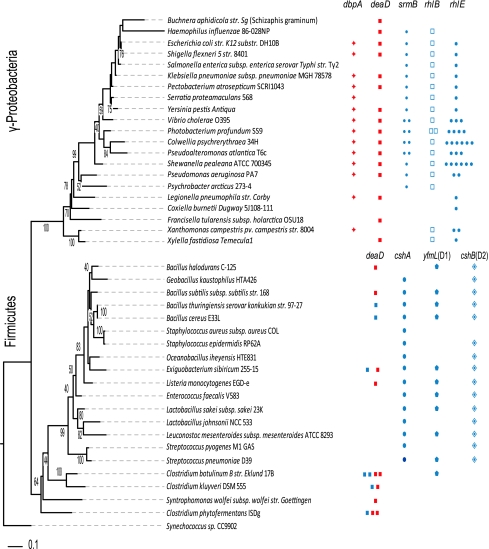
Fig. 4.
Relationship between phylogeny and the presence of putative ortholog sets of DEAD box proteins. The phylogenetic tree was based on the maximum likelihood method with the 16S rRNA gene (sequences from the RDP database) using the PhyML model (GTR + I) with 1,000 bootstrap replicates. The rows represent reference DEAD-box protein genes from E. coli (for γ-Proteobacteria) and from B. subtilis (for Firmicutes). The columns represent protein conservation using symbols to denote the presence of a putative ortholog. Red symbols are used for DEAD-box proteins containing the DbpA RNA-binding domain (RBD), while blue symbols are used for DEAD-box proteins lacking the DbpA domain. The conservation data were obtained from the phylogenetic reconstruction shown in Figs. 3 and 5, which is only based on the DEAD-Helicase_C N-terminal domain. D1 and D2 refer to the duplications 1 and 2 observed for the DEAD-box proteins from the Firmicutes. For each putative ortholog set, the length size difference is generally less than 20%, with a few exceptions (see Supplemental Tables S10 and S11 for the identification number of each protein as well as a comparison of the length for each of the sequences shown in this figure)