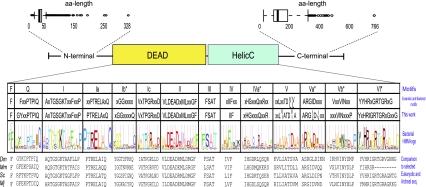
Fig. 6.
Conservation of motifs across DEAD and Helicase_C domains of 1208 bacterial DEAD-box proteins. Revised motifs obtained from the alignment of 1,208 sequences in comparison with the consensus motifs from DEAD-box proteins originally proposed by Rocak and Linder (2004) and Jankowsky and Putnam (2010). A traditional consensus residue format and a HMM-logo description are shown. The N- and C-terminal amino acid length distribution is also shown. Included for comparison are the following selected eukaryotic DEAD-box protein sequences: Dm: Drosophila melanogaster (NP_723899.1); Mm: eIF4AII Mus musculus (NP_001116510.1); Sc: Ded1 Saccharomyces cerevisiae (NP_014847.1); and Mj: Methanocaldococcus jannaschii DSM 2661(NP_247653.1)