Figure 1.
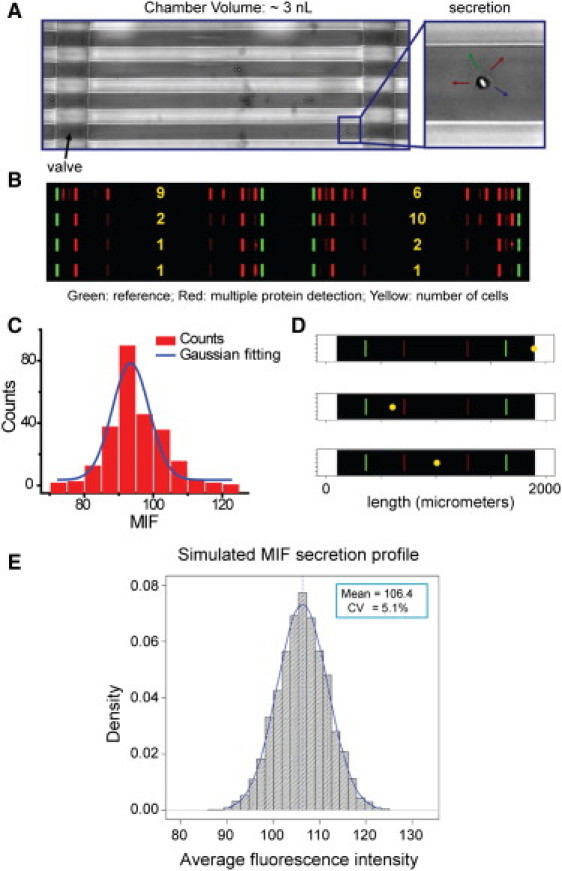
Single cell barcode chips for protein secretion profiling and experimental and simulation results for extracting the experimental error contribution to the SCBC protein assays. (A) Optical micrograph showing macrophage cells loaded into microchambers. (B) Scanned image showing the result of secretion profiling from small numbers of cells. The area flanked by two green bars corresponds to a microchamber, each of which contains two full barcodes. Each barcode represents the whole panel of assayed proteins. The barcode fluorescence image has been uniformly contrast enhanced to highlight the detected proteins. (C) Representative histogram of signal measured from individual barcode stripes for assaying a 5 ng/ml solution of recombinant MIF protein, representing a Gaussian distribution with a coefficient of variation (CV) near 7%. (D) Monte-Carlo simulated barcode intensity (corresponding to MIF) versus cell location in three single cell chambers. Yellow dots represent cell locations, and the brightness of the red stripes reflects the simulated signal level. The cell-location effect is minimized by averaging the signals from both barcodes. (E) Histogram from simulations of 5000 single cell experiments. For this simulation, the diffusion equation was solved with a randomly located, continuously secreting cell. The histogram represents the averaged intensities over both barcodes, and includes the experimentally determined barcode variability.
