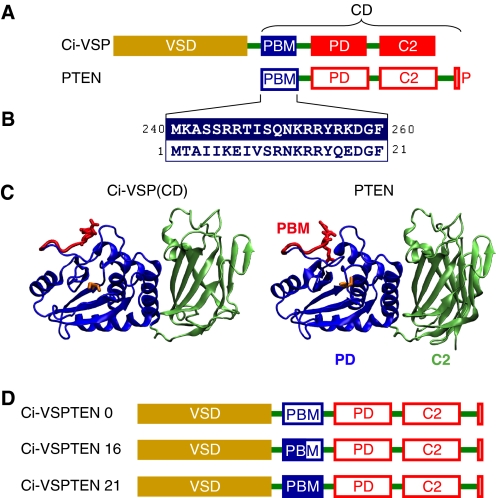
FIGURE 1.
Design of chimeras between Ci-VSP and PTEN. A, comparison of the domain organization of Ci-VSP and PTEN. P, PDZ-binding motif. B, high sequence identity between the PBMs of Ci-VSP and PTEN. Numbers refer to the amino acid positions in Ci-VSP and PTEN, respectively. C, structural similarity between the catalytic domains of Ci-VSP and PTEN predicted by structural homology modeling of the CD of Ci-VSP (left panel) based on the crystal structure of PTEN (26), generated with ESyPred3D (48). PD and C2 domains are displayed in blue and green, respectively. Partially rendered PBMs are shown in red, with arginines 253 and 254 in Ci-VSP and arginines 14 and 15 in PTEN emphasized as stick models. Cysteines 363 (in Ci-VSP) and 124 (in PTEN) in the catalytic core are displayed as yellow stick models. D, design of Ci-VSPTEN chimeras. PTEN replaces the CD of Ci-VSP with different variants of the PBM (see text).