Abstract
Enveloped viruses must fuse the viral and cellular membranes to enter the cell. Understanding how viral fusion proteins mediate entry will provide valuable information for antiviral intervention to combat associated disease. The avian sarcoma and leukosis virus envelope glycoproteins, trimers composed of surface (SU) and transmembrane heterodimers, break the fusion process into several steps. First, interactions between SU and a cell surface receptor at neutral pH trigger an initial conformational change in the viral glycoprotein trimer followed by exposure to low pH enabling additional conformational changes to complete the fusion of the viral and cellular membranes. Here, we describe the structural characterization of the extracellular region of the subgroup A avian sarcoma and leukosis viruses envelope glycoproteins, SUATM129 produced in chicken DF-1 cells. We developed a simple, automated method for acquiring high resolution mass spectrometry data using electron capture dissociation conditions that preferentially cleave the disulfide bond more readily than the peptide backbone amide bonds that enabled the identification of disulfide-linked peptides. Seven of nine disulfide bonds were definitively assigned; the remaining two bonds were assigned to an adjacent pair of cysteine residues. The first cysteine of surface and the last cysteine of the transmembrane form a disulfide bond linking the heterodimer. The surface glycoprotein contains a free cysteine at residue 38 previously reported to be critical for virus entry. Eleven of 13 possible SUATM129 N-linked glycosylation sites were modified with carbohydrate. This study demonstrates the utility of this simple yet powerful method for assigning disulfide bonds in a complex glycoprotein.
Keywords: Disulfide, Glycoprotein Structure, Glycosylation, Mass Spectrometry (MS), Retrovirus, Virus Entry
Introduction
To enter cells and begin replication, enveloped viruses must fuse the membrane coating the viral particle with a cellular membrane to deliver a subviral particle inside the cell (for review, see Refs. 1, 2). The fusion of two membranes is thermodynamically favored but comes with a very high kinetic barrier(s). Enveloped viruses have one or more glycoproteins to mediate the fusion process using the energy liberated upon conformational changes of the viral glycoprotein(s) to clear the kinetic barrier(s). Viral entry begins when the viral glycoproteins bind an appropriate cell surface protein initiating one of three possible mechanistic fates. The viral glycoprotein-receptor interaction can serve to enable trafficking of the virion into the endocytic pathway where the low pH environment triggers a conformation change in the viral glycoproteins initiating fusion of the viral and endosome membranes (e.g. influenza virus). With other viruses, for example most retroviruses, the viral glycoprotein-receptor interaction itself triggers a conformational change in the viral glycoproteins at neutral pH at the cell surface sufficient for the ultimate fusion of the viral and cellular membranes. In a third mechanism, the interaction between the viral glycoprotein and receptor at neutral pH at the cell surface triggers an initial conformational change in the viral glycoproteins. The triggered glycoproteins then require exposure to low pH to complete the conformational changes for fusion of the viral and cellular membranes (e.g. avian sarcoma and leukosis viruses (ASLVs)).3 Understanding how viral fusion proteins mediate entry will provide valuable information for antiviral intervention to combat associated disease. We are using the homologous group of retroviruses, subgroups A to E ASLV, to study enveloped virus entry because these viruses have evolved from a common ancestor to use different cellular proteins as receptors (3, 4) and have maintained the fusion process to efficiently enter cells (5–7).
All retroviral glycoproteins are synthesized as polyprotein precursors consisting of the surface glycoprotein (SU) that contains the domains that bind the cellular receptor and the transmembrane glycoprotein (TM) that anchors the protein to the membrane and contains the domains responsible for the fusion process (for review, see Ref. 8). After synthesis, the precursor polyproteins are glycosylated and form trimers through noncovalent interactions between extracellular regions of the TM glycoprotein. The viral glycoprotein precursors form a mature, metastable complex capable of mediating virus entry into the cell only after the SU and TM domains are cleaved by a cellular protease to yield a trimer of SU/TM heterodimers (9). The interaction of multiple, noncontiguous regions of SU and a specific receptor protein are required to initiate the entry process by triggering a conformational change in the SU glycoproteins, allowing the kinetically trapped TM glycoprotein structure to extend and form a lower energy structure that projects the fusion peptide toward the target membrane. Two domains in TM are critical for the extension and the subsequent refolding of TM: the N-terminal and C-terminal heptad repeats. The lowest energy form of the TM trimer, the six-helix bundle, forms when the C-terminal heptad repeats fold back into grooves created by the N-terminal heptad repeats, bringing the viral and cellular membranes into close proximity. Fusion of the membranes goes through an initial outer lipid leaflet mixing, then hemifusion, initial pore formation, pore widening, and the completion of fusion where the retroviral glycoprotein 6HB may undergo additional structural rearrangements (10–12). The cooperation of several glycoprotein-receptor interactions is likely required to form a local environment capable of fusion.
The subgroup A to E ASLV envelope glycoproteins are highly related: differences in the hypervariable domains of SU (hr1, hr2, and vr3) define receptor usage and suggest that these viruses have evolved from a common ancestor (13). Tva, the receptor for subgroup A ASLV, is related to the ligand-binding repeat of the low density lipoprotein receptor family (14, 15). Tvb, the receptors for the subgroup B, D, and E ASLVs, is a member of the tumor necrosis factor receptor family (16–18). Tvc, the receptors for subgroup C ASLVs, is related to yet a third disparate family of proteins, the immunoglobulin Ig proteins (19). The normal physiological functions of Tva, Tvb, and Tvc are currently unknown. Engineered secreted forms of these ASLV receptors, the extracellular domain of the receptor alone or fused to a IgG domain, retain biological activity sufficient to bind the viral glycoproteins with high affinity and trigger conformational changes in the viral glycoproteins similar to changes expected to occur during initiation of the infection process (20–22). Engineered secreted forms of the SU glycoproteins either alone or fused to an IgG domain can bind the soluble forms of the receptors in a subgroup-specific manner (23). The secreted forms of the ASLV SU or its receptor act as competitive inhibitors, having a significant antiviral effect on the specific ASLV subgroup infection of cells (22, 24).
We are characterizing the structural and functional organization of the ASLV glycoproteins to understand better how the ASLV fusion proteins mediate the entry process (25). Because the number and location of the cysteine residues, 14 Cys in SU, 5 in TM, and 13 of 14 possible N-linked glycosylation sites, 12 in SU, 2 in TM, are conserved in the subgroup A to E ASLV glycoproteins, it is likely these glycoproteins share a common structure and glycosylation pattern. Unusual for retroviruses, the ASLV TM glycoprotein has an internal fusion peptide 21 residues downstream of the N terminus with 2 cysteine residues flanking the fusion peptide. The organization of the functional domains of the ASLV TM fusion protein is remarkably similar to the Ebola virus fusion protein, GP2, although the ASLV SU and Ebola virus GP1 glycoproteins are not (26, 27). A disulfide bond reported to be stable throughout the fusion process covalently connects the ASLV SU and TM glycoproteins. If we assume that the disulfide bond links 1 cysteine from SU and 1 cysteine from TM, then there is at least 1 cysteine in SU that is an unbound, free cysteine. Recently, Smith and Cunningham reported that the formation of a reactive cysteine thiolate at residue Cys38 in the subgroup A SU glycoprotein was absolutely required for productive ASLV infection but was not necessary for the glycoprotein binding of the Tva receptor or Tva-triggered conformational changes (28). Also, there was no evidence that the cysteine thiolate mediated isomerization of the SU-TM disulfide bond.
In this study we physically mapped the secondary structure of an engineered secreted form of the subgroup A ASLV envelope glycoprotein (Fig. 1A). Einfeld and Hunter describe a secreted form of the subgroup A envelope glycoprotein truncated at TM residue 129 (SUATM129) (Fig. 1B), deleting the transmembrane and cytoplasmic domains, which was still capable of forming oligomers (likely trimers) detected by sucrose gradient (29). The SUATM129 glycoprotein was expressed in chicken DF-1 cells (30, 31), a natural cell substrate for ASLV infection and replication and highly purified from cell culture supernatants for this study. We used this biologically active, complex glycoprotein to develop a simple, automated method for acquiring high resolution mass spectrometry data that enabled the identification of disulfide-linked peptides, the assignment of most of the disulfide bonds, and the identification of the sites of N-linked glycosylation from a single HPLC run.
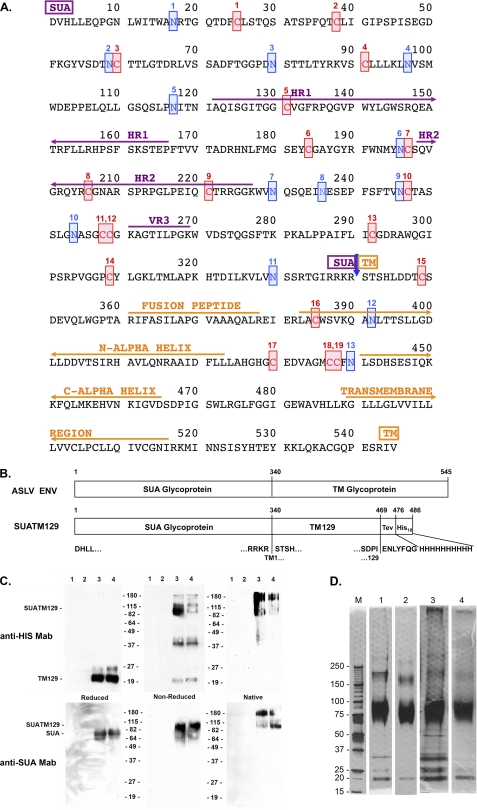
FIGURE 1.
Amino acid sequence of the mature subgroup A ASLV envelope glycoprotein and analysis of the purified SUATM129 glycoprotein. A, functional regions of SUA, hr1, hr2, and vr3 primarily responsible for interacting with the Tva receptor, TM, the internal fusion peptide domain, the N- and C-α helices and the transmembrane domain, are indicated. The 19 cysteine residues are numbered and highlighted in red. The 13 possible N-linked glycosylation sites are numbered and highlighted in blue. SUA and TM are cleaved after amino acid 340 (blue arrow). B, schematic representations of the mature wild-type ASLV(A) and SUATM129 envelope glycoproteins. The TEV protease cleavage site and the His10 tag are shown fused to TM residue 129. C, Western immunoblot analysis of SUATM129 purified from two negative cell lines (lanes 1 and 2) and two cell lines expressing SUATM129 (lanes 3 and 4). Samples were either left nonreduced or reduced with β-mercaptoethanol and separated by SDS-PAGE, or left in the native state and separated by PAGE using Criterion 4–15% gradient gels. The proteins were transferred to nitrocellulose and the Western immunoblots probed first with either an anti-His or anti-SUA monoclonal antibody, and then a peroxidase-conjugated goat anti-mouse IgG. The bound complexes were visualized by chemiluminescence. Molecular mass markers are in kDa. D, SDS-PAGE analysis of two different purified SUATM129 preparations (preparation 1 in lanes 1 and 2; preparation 2 in lanes 3 and 4). Nonreduced SUATM129 samples after metal affinity purification (lanes 1 and 3) or after metal affinity plus octyl-Sepharose purification (lanes 2 and 4) were separated by SDS-PAGE using Criterion 4–15% gradient gels. The proteins were visualized with silver stain. Molecular mass markers (lane M) are in kDa.
EXPERIMENTAL PROCEDURES
Materials
All of the proteases used, trypsin, chymotrypsin, and Asp-N- endoproteinase, were sequencing grade and purchased from Roche Diagnostics. The PNGase F was purchased from New England Biolabs, and the Zwittergent 3-16 from EMD Chemicals (Gibbstown, NJ). The solvents were purchased from Honeywell Burdick and Jackson (Morristown, NJ), and the trifluoroacetic acid (TFA) and formic acid were purchased from Fluka. The tris (2-carboxyethyl) phosphine (TCEP) and N-ethylmorpholine were purchased from Sigma-Aldrich.
Cloning and Expression of the SUATM129 Glycoprotein
To aid in the purification of the SUATM129 glycoprotein, a 10-histidine residue tag preceded by the TEV protease cleavage sequence was added in-frame after amino acid 469 of the SR-A enveloped protein (Fig. 1). An adapter plasmid encoding the SUA-rIgG immunoadhesin, PUCCLA112N-SUA-rIgG, was described previously (23). The IgG region was replaced with the SalI to MfeI fragment of the SR-A env gene yielding a coding region that begins with a consensus Kozac start site, NcoI site, and encodes the complete leader region and the SR-A envelope to residue 469. Oligonucleotides were then used to add the TEV protease and His10 sequences by cloning as an MfeI to PstI fragment into PUCCLA112-SUATM129. The complete and tagged SUATM129 expression cassette was isolated as a ClaI fragment and cloned into the TFANEO expression plasmid under the transcriptional control of SR-A LTRs (32). TFANEO also encodes a neo expression cassette for selection.
DF-1 cells (30, 31) were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 100 units of penicillin/ml, and 100 μg of streptomycin/ml (Quality Biological, Gaithersburg, MD) at 39 °C and 5% CO2. DF-1 cells transfected with the TFANEO-SUATM129 plasmid were grown in 500 μg of G418/ml to select for neomycin-resistant cells. Clones were isolated using cloning cylinders (Bellco Glass Inc., Vineland, NJ), expanded, and maintained with standard medium supplemented with 250 μg/ml G418.
Purification of the SUATM129 Glycoprotein
When the SUATM129-expressing cells were confluent, the medium was changed to a serum-free medium, VP-SFM (Invitrogen) supplemented with 4 mm l-glutamine (Invitrogen), harvested 24 h later, and cleared of debris using a 3 μm capsule filter (12116; PALL). The cleared supernatant was mixed with TALON Metal Affinity Resin (Clontech), prewashed, and equilibrated with wash buffer (50 mm sodium phosphate, pH 7.0, 300 mm NaCl) for 90 min at room temperature to bind the SUATM129 protein His10 tag, and then loaded into a chromatography column by gravity flow. The resin was washed with wash buffer (500 bed volumes) and then washed with 2.5 mm imidazole followed by 10 mm imidazole in wash buffer (100 bed volumes each). The resin was then eluted first with 5 bed volumes of 75 mm imidazole followed by 5 bed volumes of 250 mm imidazole in wash buffer, and the fractions were combined, concentrated and the buffer exchanged with 100 mm sodium phosphate, pH 7.0, using JumboSep concentrators with 10K filters (PALL). To purify the SUATM129 protein further from contaminating proteins, ammonium sulfate was added to the preparation, resulting in a loading buffer of 1 m ammonium sulfate, 100 mm sodium phosphate, pH 7.0, and loaded onto an octyl-Sepharose column. The column was washed with a gradient exchanging the load buffer to 100 mm sodium phosphate, pH 7.0, buffer. The SUATM129 protein was eluted first with water, recovering ∼60–70% of the protein, followed by elution with 20% ethanol that recovered some additional SUATM129 protein. Fractions containing SUATM129 protein were combined and concentrated, and the buffer was exchanged with 10 mm HEPES, pH 7.4, 100 mm NaCl, 5% glycerol for storage.
PAGE and Western Transfer Analysis
To analyze reduced proteins, the samples were adjusted to 1× Laemmli loading buffer (2% SDS, 10% glycerol, 50 mm Tris-Cl, pH 6.8, 5% β-mercaptoethanol, 0.1% bromphenol blue) and boiled for 5 min. To analyze nonreduced proteins, the samples were adjusted to 1× Laemmli loading buffer without the β-mercaptoethanol (2% SDS, 10% glycerol, 50 mm Tris-Cl, pH 6.8, 0.1% bromphenol blue) and boiled for 5 min. To analyze native proteins, the samples were adjusted to 1× Laemmli loading buffer without the SDS and β-mercaptoethanol (10% glycerol, 50 mm Tris-Cl, pH 6.8, 0.1% bromphenol blue) and loaded directly. Proteins were separated on Criterion 4–15% Tris-HCl gradient polyacrylamide gels (Bio-Rad) and either silver stained directly (33), or the proteins were transferred onto nitrocellulose membranes.
The Western transfer filters were blocked in phosphate-buffered saline (PBS) with 10% nonfat dry milk for 1 h at 25 °C. The filters were then rinsed briefly in rinse buffer (100 mm NaCl, 10 mm Tris-Cl, pH 8, 1 mm EDTA, 0.1% Tween 20) and incubated with either an unconjugated anti-His antibody (1:3000 dilution) (27471001; GE Healthcare) or anti-ASLV SU(A) monoclonal antibody (34) (purified from the mc8C5 hybridoma, a kind gift from Christina Ochsenbauer-Jambor and Eric Hunter, University of Alabama at Birmingham, AL) (1:1000 dilution) in rinse buffer containing 1% nonfat dry milk for 1 h at 25 °C. The filters were washed extensively with rinse buffer and then incubated with 50 ng/ml peroxidase-labeled goat anti-mouse IgG (H+L) (Kirkegaard and Perry, Gaithersburg, MD) in rinse buffer with 1% nonfat dry milk for 1 h at 25 °C. After extensive washing with rinse buffer, immunodetection of the protein-antibody-peroxidase complexes was performed with Western blotting Chemiluminescence Reagent (PerkinElmer Life Sciences). The immunoblots were then exposed to Kodak X-Omat film.
ALV Alkaline Phosphatase Assay
For alkaline phosphatase assays, DF-1 cell cultures (∼30% confluent) were incubated with 10-fold serial dilutions of the appropriate RCASBP/alkaline phosphatase virus stocks for 36–48 h. The assay for alkaline phosphatase activity was described previously (22).
Protease Digestions and HPLC
Trypsin digestions were done as follows. 100 μg of SUATM129 was initially digested with 2.5 μg of trypsin in 100 mm Tris, pH 8.4, 0.004% Zwittergent 3-16 buffer at 55 °C for 2 h. An additional 2.5 μg of trypsin in 100 mm Tris, pH 8.4, 0.004% Zwittergent 3-16 buffer was then added to the reaction and incubated at 37 °C overnight. Chymotrypsin digestions were done as follows. 100 μg of purified SUATM was digested with 5 μg of chymotrypsin in 100 mm Tris, pH 8.5, 0.004% Zwittergent 3-16 buffer at room temperature overnight. To remove all N-linked glycosylation, PNGase F (500 units/μl) (3000 units) was added to the postdigest reactions and incubated at 37 °C for 3 h. Each digest mixture was split into two aliquots, with the disulfide bonds reduced in one aliquot by adding TCEP (25 mm final) and incubating at 55 °C for 2 h.
The nonreduced or reduced peptide mixtures were each separated with a Zorbax 300 SB-C18, 5-μm, 150 × 0.5-mm reverse phase column using an Agilent 1100 Series Capillary HPLC system at a flow rate of 15 μl/min and collecting 1-min fractions using gradients described in Table 1. Detection was by UV absorbance at 214 nm. All fractions were then dried down using a SpeedVac spinning concentrator and stored at −80 °C. Fractions were reconstituted with 40% acetonitrile, 5% isopropyl alcohol, and 0.2% formic acid prior to analysis by direct chip-based infusion Fourier Transform ion cyclotron resonance mass spectrometry using an LTQ-FTICR-MS with electron capture dissociation (ECD) and an Advion Nanomate 100.
TABLE 1.
HPLC running conditions for peptide separation
| Protease | Gradient | Time | % B |
|---|---|---|---|
| min | |||
| SUATM/trypsin | Phase A: 5% acetonitrile/0.1% TFA | ||
| Phase B: 80% acetonitrile/0.1% TFA | 0 | 5 | |
| 15 | 5 | ||
| 115 | 45 | ||
| 117 | 80 | ||
| 120 | 80 | ||
| 122 | 5 | ||
| 135 | 5 | ||
| SUATM/chymotripsin | Phase A: 5% acetonitrile/0.1% TFA | ||
| Phase B: 80% acetonitrile/0.1% TFA | 0 | 2 | |
| 5 | 2 | ||
| 10 | 8 | ||
| 59 | 30 | ||
| 69 | 80 | ||
| 72 | 80 | ||
| 73 | 2 | ||
| 78 | 2 |
Endoproteinase Asp-N was utilized for the fractions found to contain adjacent cysteines. The digests were performed on dried HPLC fractions that were resolubilized in 100 mm Tris, pH 8.4, with 4 ng of Asp-N and digested at 37 °C overnight prior to analysis by nanoLC-ESI-MS/MS. The fractions that were analyzed by chip-based direct infusion FTICR-ECD-MS were redried and solubilized in 0.2% N-ethylmorpholine buffer plus 1 ng of Asp-N and digested at 37 °C for 1.5 h.
Chip-based Infusion-FTICR-MS with ECD
The HPLC fractions were resolubilized in 30 μl of 40% acetonitrile, 0.2% formic acid, 5% isopropyl alcohol. All disulfide-linked peptides were identified by screening the fractions with chip-based ESI infusion using an Advion Nanomate 100 coupled to a ThermoFinnigan LTQ-FT Hybrid Ion Cyclotron Resonance Mass Spectrometer upgraded with the ECD capability. The Nanomate was set to pick up 6 μl of sample and spray at 1.5 kV with N2 back-pressure of 0.3 pound/square inch. Each fraction was screened for 3 min, performing data-dependent acquisitions consisting of a FT full scan from 300 to 2000 m/z with the resolving power set at 100,000 @ 400 m/z followed by an ECD scan from 100–2000 m/z (3 microscans, 100,000 R, energy = 4.0, delay = 10 ms, duration = 100 ms) on ions with charge states of [M+2H]2+ or higher, plus a linear ion trap collision-induced dissociation (CID) scan on ions with charge states of [M+2H]2+ and [M+3H]3+. Each ion was analyzed twice then placed on an exclusion list for 60 s. All ECD spectra were analyzed manually by looking for the presence of theoretical trypsin-cleaved peptide ions. Non-cysteine-containing peptide ions were identified from analysis of the CID-MS/MS spectra. ThermoFinnigan Xcalibur QualBrowser software was used to determine the theoretical isotopic distributions for the isotope clusters.
Orbitrap CID Fragmentation
Fractions containing peptides with adjacent cysteines were analyzed by nanoLC-ESI-MS/MS using a ThermoFinnigan LTQ Orbitrap Hybrid Mass Spectrometer (Thermo Fisher Scientific) coupled to a nanoLC-two-dimensional HPLC system (Eksigent, Dublin, CA). The chymotrypsin fraction containing the C1 and C17C18C19 peptides was treated with Asp-N protease, after which an aliquot was diluted with 0.15% formic acid and 0.05% TFA and loaded onto a 250-nl OPTI-PAK trap (Optimize Technologies, Oregon City, OR) custom packed with Michrom Magic C8 solid phase (Michrom Bioresources, Auburn, CA). Chromatography was performed using 0.2% formic acid in both the A solvent (98% water and 2% acetonitrile) and B solvent (80% acetonitrile, 10% isopropyl alcohol, and 10% water), and a 10% B to 50% B gradient over 45 min at 325 nl/min through a hand-packed PicoFrit (New Objective, Woburn, MA) nanobore (Michrom Magic C18 3-μm 75 μm × 200 mm) column. The LTQ Orbitrap mass spectrometer experiment consisted of a FT full scan from 300 to 1200 m/z with resolution set at 60,000 (at 400 m/z), followed by Orbitrap CID MS/MS scans on the top three ions at 30,000 resolution. Dynamic exclusion was set to 2, and selected ions were placed on an exclusion list for 30 s. The lock-mass option was enabled for the FT full scans using the ambient air polydimethylcyclosiloxane ion of m/z = 445.120024 or a common phthalate ion m/z = 391.284286 for real-time internal calibration (35). The MS/MS spectra ions were assigned manually.
Edman Degradation N-terminal Chemical Sequencing
Relative abundance of the disulfide-linked peptides was determined for some of the HPLC fractions using Edman degradation N-terminal chemical sequencing on a Procise cLC Protein Sequencer (Applied Biosystems, Foster City, CA). The amino acid assignments were made manually, and the relative amounts of each peptide were determined from the amino acid yields of the first few sequencing cycles.
RESULTS
SUATM129 Protein
The SUATM129 glycoprotein was produced in chicken cells, normal host cells for ASLV replication, to ensure appropriate envelope glycoprotein folding, glycosylation, and transport. DF-1 cell lines were established that stably produced SUATM129. SUATM129 was expressed as a secreted protein with a histidine tag to allow a relatively simple purification procedure of biologically active protein from cell culture supernatants in good yield (see “Experimental Procedures”). Biological activity was demonstrated by the ability of the SUATM129 protein to bind a soluble form of the ASLV subgroup A Tva receptor as shown by immunoprecipitation (data not shown) and the ability of SUATM129 to inhibit cell surface expressed Tva and specifically block ASLV(A) infection of normally susceptible cells by greater than 10–50-fold compared with another ASLV subgroup (data not shown). The calculated molecular mass of SUATM129 is 53,496 Da; the SUA subunit (37,265 Da) connected to the TM129 subunit (16,249 Da) by a disulfide bond. Previously published work on the secreted SUA-rIgG immunoadhesin, the complete SUA subunit fused to a rabbit IgG domain, identified that 10 of 11 potential N-linked glycosylation sites in SUA were glycosylated with ∼23 kDa of carbohydrate. If the SUATM129 glycoprotein contains similar levels of glycosylation, the SUATM129 glycosylated molecular mass would be at least 76,496 Da; the SU subunit ∼60,264 Da. The ASLV TM subunit has two possible sites of N-linked glycosylation that could add additional carbohydrate to the SUATM129 glycoprotein.
The SUATM129 expressed by cell lines was initially characterized by small scale purification using a nickel affinity resin to bind the His tag and the eluted proteins analyzed by PAGE and Western immunoblots using an antibody against the HIS-tag and an antibody that specifically binds the SUA subunit. Proteins were either reduced or left nonreduced before SDS-PAGE to break or maintain the disulfide bond linking SUA and TM, respectively, or the proteins were left in the native state and separated by PAGE. Fig. 1C is an example of this analysis. The nonreduced SUATM129 protein migrated at ∼85 kDa, with the reduced protein yielding the SUA subunit at ∼75–80 kDa and the TM129 at ∼20 kDa. The native gel showed that at least some of the SUATM129 glycoprotein was migrating as an oligomeric form or forms as predicted from the initial report on this protein. Large scale purification of SUATM129 yielded a protein mixture predominantly consisting of SUATM129 but also containing several other proteins (Fig. 1D, lanes 1 and 3). Analysis of the protein bands from the SDS-PAGE separated nickel-resin-purified protein preparation by mass spectrometry identified fibronectin, lactate dehydrogenase, and lamin A as the major contaminants (data not shown). This initial purified product was further purified using octyl-Sepharose chromatography that improved the overall purity of the final SUATM129 product (Fig. 1D, lanes 2 and 4). The SUATM129 glycoprotein preparation purified by metal affinity chromatography followed by octyl-Sepharose chromatography was used for all subsequent experiments.
HPLC Analysis of SUATM129 Peptides
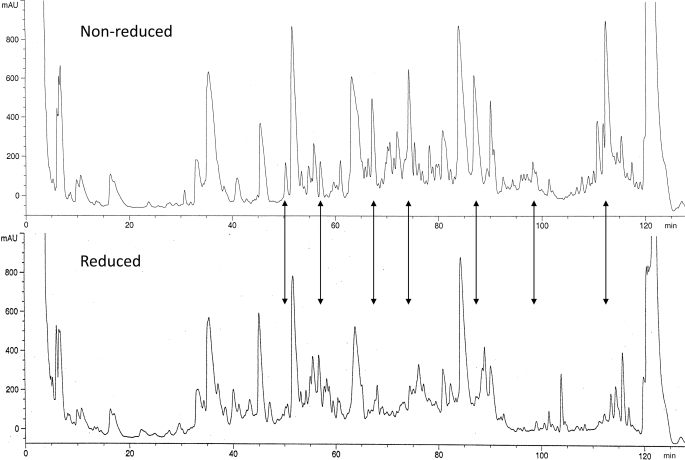
The SUATM129 glycoprotein presented several challenges for enzymatic digestion and HPLC of the peptides due to carbohydrate content and the desire to maintain the disulfide bonds. Attempts to remove the carbohydrate from intact SUATM129 prior to enzymatic digestion resulted in the precipitation of the deglycosylated protein that was difficult to resolubilize. Subsequent enzymatic digests resulted in chromatograms with inconsistent, poor peak shapes (data not shown). Better chromatography performance was observed when SUATM129 was first digested into peptides with a protease and then deglycosylated with PNGase F. The upper panel in Fig. 2 shows the HPLC chromatogram of SUATM129 following trypsin digestion and deglycosylation, with the disulfide bonds intact. Half of the same digest mixture was then reduced with TCEP and run under identical HPLC conditions (lower panel). The chromatogram for the nonreduced mixture clearly shows peaks that are missing from the chromatogram of the disulfide-reduced digest mixture. It is not clear in the TCEP-treated chromatogram where the reduced cysteine peptides are eluting. The fractions corresponding to the nonreduced peaks contain disulfide-linked peptides that were initially used to develop the automated chip-based direct infusion electrospray FTICR method on the LTQ-FT. Once the methods were established, all HPLC fractions from the protease digests were screened and analyzed.
FIGURE 2.
Reverse-phase HPLC chromatograms of nonreduced or reduced, trypsin-digested and deglycoslyated SUATM129 peptides. Some of the peptide peak changes between the nonreduced and reduced chromatograms, presumably due to the breaking of a disulfide bonded peptide pair, are highlighted with arrows.
FTMS Analysis of SUATM129 Peptide Pairs
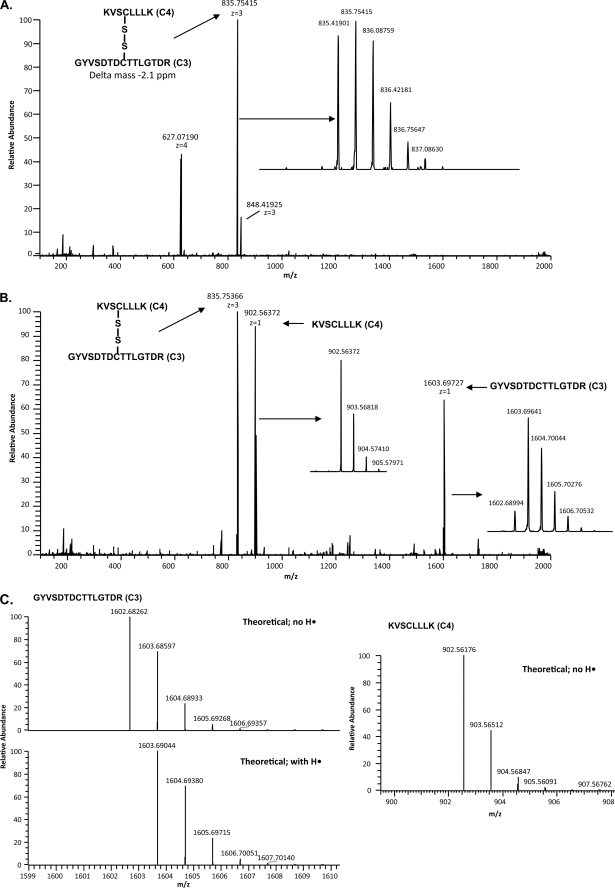
Previous studies have shown that performing ECD on disulfide-paired peptides cleaves the disulfide bond more readily than the peptide backbone amide bonds (36, 37). The LTQ-FT with the ECD option is a hybrid mass spectrometer that uses the linear ion trap for the controlled isolation of an ion and the FTICR cell to perform ECD on that ion, scanning the resulting fragments with high mass accuracy. In this study, we used chip-based infusion with the Advion Nanomate on an LTQ-FT to develop ECD conditions that minimized peptide bond fragmentation and give ECD fragment spectra containing both the intact disulfide-paired ion and the disulfide-cleaved peptide ions. These conditions were used in the LTQ-FT data-dependent method to screen the HPLC fractions from the nonreduced SUATM129 protease digests. The method provided accurate full scan mass data and accurate mass ECD spectra giving predominantly the parent ion corresponding to the mass of the disulfide linked peptide pair and the ions of the dissociated peptides as fragment ions. The ions were manually compared with the theoretical ions corresponding to protease-specific SUATM129 cysteine-containing peptides. The FT full scan and the ECD spectrum in Fig. 3 illustrate the method used to assign the C3 and C4 peptides as a disulfide-bonded pair. The C3+C4 ion 835.41901 [M+3H]3+ was isolated (Fig. 3A) and cleaved with ECD, and the resulting ions were analyzed identifying the original C3+C4 ion, the C3 ion 1602.68994 [M+H]1+, and the C4 ion 902.56372 [M]•+ (Fig. 3B). The mechanism of ECD disulfide bond cleavage often results in one of the peptides acquiring a hydrogen atom on the cysteine sulfur while the other peptide cysteine sulfur does not (36, 37). Analysis of the ECD-cleaved C3 and C4 ions shows an isotopic distribution of the singly charged C4 peptide ion with a cysteine sulfur that did not acquire a hydrogen, whereas the C3 peptide ion was protonated, though not completely (Fig. 3C).
FIGURE 3.
Example of the mass spectrometry analysis of a disulfide-bonded peptide pair using ECD. A, FTICR full scan of the HPLC separation of trypsin-digested SUATM129 protein fraction 57 is shown. The ion with an exact mass corresponding to the C3 peptide and the C4 peptide connected by a disulfide bond is indicated. The isotopic distribution of the peptides is shown in the inset. B, ECD scan triggered from the 835.41901 [M+3H]3+ ion is shown. Experimental conditions were used so that each of the singly charged ECD fragment ions as well as the parent ion were present in the ECD spectra. The singly charged ECD fragment ions add up to the mass of the parent ion. The isotopic distributions of the ECD fragment peptides are shown in the insets. C, ECD fragmentation can produce peptides with variable protonation. The isotopic distribution of the ECD fragment peptides shows that the resulting cysteine sulfurs can be partially protonated (C3) or completely deprotonated (C4) compared with the theoretical distributions.
A summary of the FTICR-ECD analyses of the HPLC fractions containing the nonreduced digest peaks is shown in Fig. 4. The differences between the observed (measured) peptide masses, both parent ion and ECD-generated ions and the theoretical (calculated) masses, are reported as delta mass in parts per million (35). Four fractions from the nonreduced trypsin digest were found to contain a single peptide pair with the ECD spectra demonstrating that C3 is bound to C4 in fraction 57, C7 is bound to C8 in fraction 50, C13 is bound to C14 in fraction 87, and C15 is bound to C16 in fraction 67 (data not shown).
FIGURE 4.
Summary of the MS analysis of disulfide-bonded peptide pairs of SUATM129. The accurate mass measurements of the disulfide-bonded peptide pairs were made using FTICR mass spectrometry with an LTQ-FT. The measured masses are shown as mass/ion charge (m/z). ECD was used to preferentially break the disulfide bond of the peptide pair to yield the original peptide pair, and each peptide mass was measured (e.g. ECD C3; ECD C4). One difference to note between using ECD compared with chemical reduction of a disulfide bond: ECD does not necessarily result in peptides with replaced hydrogens on all of the now free sulfur atoms. Peptides generated by ECD that have (+) or have not (−) replaced the hydrogen ions are indicated in the H+ column. For peptide pair C5+C6, the fraction was reduced and analyzed by LTQ-FTICR MS to measure the masses of the reduced peptides (redC5; redC6). Peptide pairs from CID that define the disulfide bond pattern between cysteines C1 and C17,18,19 are shown with the complete data presented in Fig. 7. The C1, C2+C17, C18, C19 peptide pair was identified only by Edman chemical sequencing. The analytical difference between the observed mass compared with the theoretical mass is given as Delta Mass in parts per million. The peptide sequences corresponding to the measured mass are shown with the putative disulfide bonds. The SUATM129 glycoprotein contains two adjacent cysteine pairs: C11,C12 and C18,C19 (marked in boxes). The methionine in the C17, C18, C19 peptide in Fraction 41 was found to be oxidized (oxM) after freeze/thaw from storage: this was the predominant form in the Asp-N and CID analysis. We consistently observed an unusual chymotrypsin cleavage between Gln29 and Ser30 generating the C1 peptide CLSTQ in multiple digests. We have no definitive explanation, but it may reflect the extended overnight digestion at room temperature.
The FT-MS full scan of fraction 112 contained a single predominant ion 1258.93970 [M+6H]6+. The ECD spectra on this ion did not produce any useful ions, in part, due to the overall low intensity of the parent ion and the large size of the C5 peptide. We hypothesized that this ion may be the C5 and C6 peptide disulfide pair based on the expected mass. Attempts using a second protease to decrease the length of the presumed C5 peptide failed to produce any clear information, so the fraction was treated with TCEP to see whether the 1259 ion responded to reduction. Another FT-MS full scan of the TCEP-treated fraction 112 yielded multiply charged ions corresponding to C5 and C6, demonstrating that C5 is bound to C6 in SUATM129. The observed peptide masses and isotopic mass distributions were all in close agreement with their theoretical masses (Fig. 4) and theoretical isotopic distributions (data not shown).
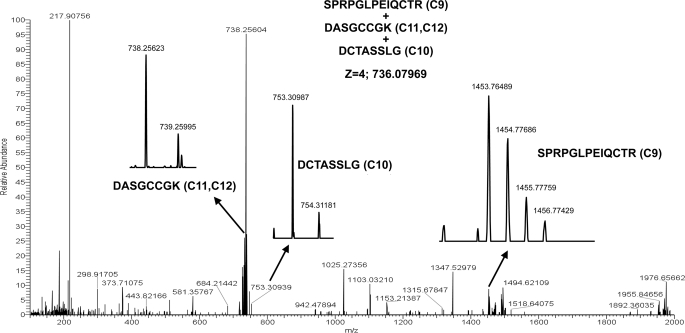
Fraction 74 contained the ion 1010.05450 [M+5H]5+, which produced an ECD spectrum with a 726.88186 [M+2H]2+• ion corresponding to the C9 peptide and a 1797.74756 [M+2H]2 + 3• ion corresponding to the C10, C11, and C12 peptide (data not shown). It is unclear from these data as to which cysteine the C9 peptide was linked although it was unlikely to be C10 because that would mean the adjacent cysteines C11 and C12 would be a disulfide-bonded pair. We noted that there was an aspartic acid between C9 and C10 and one between C10 and C11 so the remainder of that fraction was treated with Asp-N protease generating three disulfide-linked peptide chains: one peptide with C9, one with C10, and one peptide with C11C12. Subsequent MS analysis detected the ion 736.07969 [M+4H]4+ corresponding to all three peptides bonded together, ruling out C9 being disulfide-linked to C10 (data not shown). The ECD cleavage spectra of the [M+4H]4+ ion at m/z 736.07969 contained the expected masses corresponding to the C9 peptide, the C10 peptide, and the C11C12 peptide (Fig. 5). The observed peptide masses and isotopic mass distributions were in close agreement with the theoretical masses (Fig. 4) and theoretical isotopic distributions (data not shown). These data allowed us to conclude that C9 is bound to either C11 or C12 with C10 bound to the opposite cysteine.
FIGURE 5.
LTQ-FTICR ECD MS analysis of the 4+ ion of the SUATM129 trypsin+Asp-N C9+C10+C11, C12 disulfide-bonded peptides, mass 736.07969. MS analysis of the ECD reaction shows the 4+ ion mass of the original bonded peptides and each individual peptide: the peptide (z = 1) containing C9, the peptide (z = 1) containing C10, and the peptide (z = 1) containing C11 and C12.
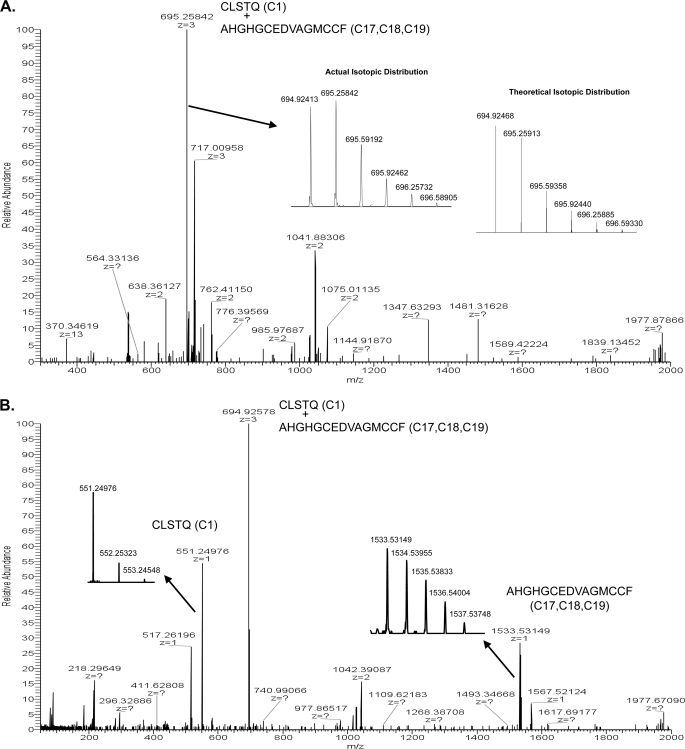
Analysis of the trypsin digest HPLC fractions did not yield any useful MS data for the assignments of the C1, C2 peptide, or the C17, C18, C19 peptide. We identified the C1, C2+C17, C18, C19 peptide pair only by Edman chemical sequencing (Fig. 4). Therefore, a chymotrypsin digest followed by PNGaseF treatment was performed on a new portion of the purified SUATM129 protein. The digest was fractionated by reverse-phase HPLC, and fractions were analyzed by ECD-MS similar to the trypsin digest. The FT full scan of chymotrypsin HPLC fraction 41 contains a 694.92413 [M+3H]3+ ion that corresponds to the theoretical mass of the C1 and C17, C18, C19 peptide pair connected with a disulfide bond (Fig. 6A). The observed mass and isotopic mass distribution were in close agreement with the theoretical mass (Δ mass −0.8 parts per million) and theoretical isotopic distribution. The spectra from ECD cleavage of the [M+3H]3+ ion contained the parent [M+3H]3+ ion plus two singly charged ions corresponding to the C1 peptide ([M+H]1+ at m/z 551.24976) and the C17C18C19 peptide ([M+H]1 + 3• at m/z 1533.53149) (Fig. 6B). The observed peptide masses and isotopic mass distributions of these peptides were in close agreement with their theoretical masses (Fig. 4) and theoretical isotopic distributions (data not shown). Similar to the C9, C10, C11, and C12 assignments discussed above, it was not clear where the C1 peptide disulfide was linked to the C17, C18, C19 peptide. Again, there is an aspartic acid between C17 and C18, and C18, C19 are adjacent cysteines. A secondary digestion of a portion of fraction 41 with Asp-N protease, followed by FT-MS analysis showed a 706.26166 [M+3H]3+ ion which corresponds to the theoretical mass of the three disulfide-linked peptides (Fig. 4; data not shown). These data indicate that C1 is not linked to C17 and must therefore be linked to C18 or C19, which suggests SUA is linked to TM through C1 and either C18 or C19, and C17 is bound to the opposite cysteine.
FIGURE 6.
LTQ-FTICR MS analysis of the SUATM129 chymotrypsin peptide pair containing cysteine C1 and cysteines C17,18,19. A, full scan of the chymotrypsin digest of SUATM129 HPLC fraction 41 is shown with accurate mass measurement of the 3+ ion (z = 3) of the C1 and C17, C18, C19 peptide pair, mass = 694.92413. The actual isotopic distribution and the theoretical isotopic distribution of mass 694.92413 are shown as insets. B, LTQ-FTICR ECD MS analysis of mass 694.92413 corresponding to the 3+ ion of the SUATM129 chymotrypsin C1 and C17, C18, C19 disulfide-bonded peptide pair and each individual peptide 1+ ion from the parent. The isotopic distributions of the ECD C1 and C17, C18, C19 peptides are shown in insets.
CID Orbitrap MS Analysis
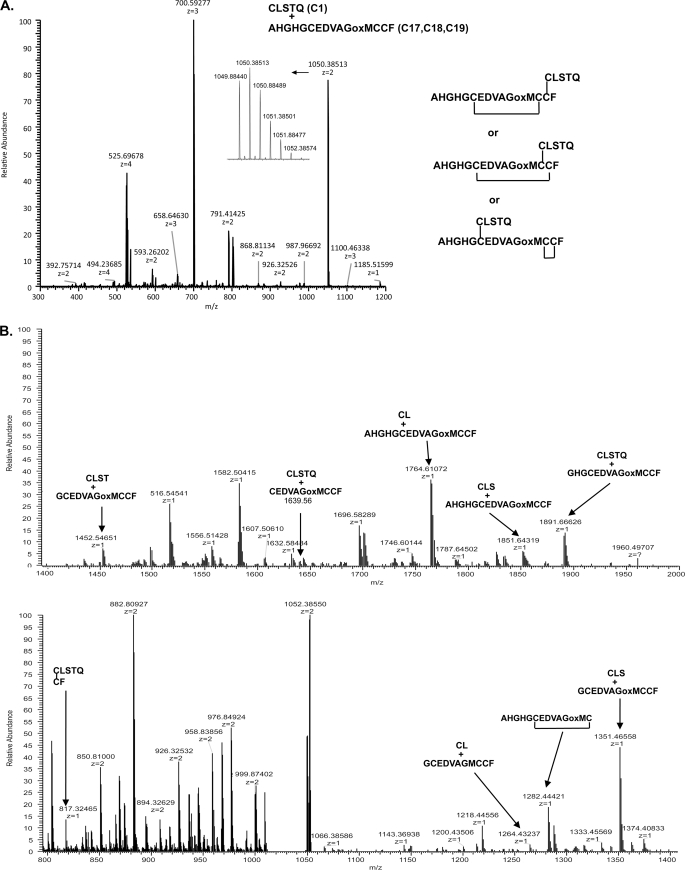
SUATM129 contains two sets of adjacent cysteine pairs, C11 and C12 in SUA, and C18 and C19 in TM (Fig. 1A). In an effort to try and assign the specific disulfide bonds to these adjacent cysteine pairs, Orbitrap CID MS/MS analysis was employed in the hope of creating accurate mass fragments generated from cleavage of the peptide bond between the adjacent cysteines that still had an intact disulfide bond. Working with a chymotrypsin fraction containing C1 and C17, C18, C19 peptides, we used the Orbitrap for the FT full scan and the analysis of the MS/MS fragments (Fig. 7A). In this experiment the methionine in the peptide pair was oxidized due to storage. The FT spectrum contains the 2+, 3+, and 4+ charge states of the disulfide-linked C1 peptide and the C17, C18, C19 peptide. The three possible disulfide pairings are also shown. The Orbitrap performed CID MS/MS on the 1049.88477 [M+2H]2+ ion; the MS/MS spectrum is shown in Fig. 7B. We generated a list of theoretical b and y ion series masses with the disulfide bonds intact and fragmentation of each peptide if the cleavage occurred between C18 and C19 and looked for those masses manually in the accurate mass MS/MS spectra (data not shown). There were many ions corresponding to fragments with C18 and C19 intact, but there was also a singly charged ion at 1282.44421 that matched a peptide fragment with C17 linked to C18. There was also a singly charged ion at 817.32465 that corresponds to peptides with C1 linked to C19 (Figs. 4 and 7B). Both of these fragments are within 4 parts per million of the theoretical fragment pair masses and are generated from a parent ion that corresponds to the C1 and C17, C18, C19 chymotrypsin peptide pair, so we propose that C1 is disulfide-linked to C19 and C17 is linked to C18. We also attempted to use this approach to assign the disulfide pairing for the C9 and the C10, C11, C12 linked peptides, but were unsuccessful: no peptides were identified resulting from cleaved between the adjacent C11 and C12 residues.
FIGURE 7.
NanoLC-Orbitrap mass spectrometry analysis of the SUATM129 chymotrypsin fraction containing the C1 and C17,18,19 disulfide-linked peptides. In this experiment the methionine in the peptide pair was oxidized (oxM) due to storage. A, FT full scan of the multiply charged ion species of the C1 and C17,18,19 peptides. All of the possible Cys-Cys bond configurations are shown. B, Orbitrap CID MS/MS spectra of the 1049.88477 [M+2H]2+ parent ion of the C1 and C17, C18, C19 disulfide-linked peptides. Some of the accurate mass fragment ion assignments including the two disulfide-linked pairs resulting from cleavage between C18 and C19 are indicated.
N-Linked Glycosylation Pattern of SUATM129
The SUATM129 protein contains 13 possible N-linked glycosylation sites (NXS/T): 11 sites in SU and 2 sites in TM (Fig. 1A). Because PNGase F treatment of an asparagine modified with carbohydrate converts the residue to an aspartic acid, the various high resolution mass spectra provided the data to assess which of these possible sites in the SUATM129 protein were in fact modified with carbohydrate. The masses and sequences corresponding to the peptides that contain a cysteine residue and a possible N-linked glycosylation site with either an unmodified asparagine or an asparagine modified with carbohydrate converted to an aspartic acid are shown in Fig. 4. Peptides containing the other possible glycosylation sites were identified in the mass spectra (data not shown). All of the MS analysis performed consistently found all possible N-linked glycosylation sites of SUATM129 to be modified with carbohydrate except sites N8 and N13 (Fig. 8). In a previous study of the related SUA-rIgG protein, a secreted protein with the SUA glycoprotein fused to a rabbit IgG domain, 10 of 11 possible N-linked glycosylations sites were modified with carbohydrate: as with the SUATM129 protein, the exception was N8 (38).
FIGURE 8.
Proposed secondary structure of the SUATM129 heterodimer of SU and TM glycoproteins. The proposed disulfide bonds are indicted with red lines. Bonds to cysteines C11 and C12 (indicated with a box) could not be assigned further. The free cysteine is indicted, C2. The N-linked glycosylation sites that were converted to aspartic acid after PNGase F treatment, and therefore contained carbohydrate additions, are marked in blue. The unglycosylated sites are underlined, N8 and N13.
DISCUSSION
We describe a novel method that enabled the assignment of multiple disulfide-linked peptides from a single HPLC separation of a nonreduced digest of a complex glycoprotein (SUATM129) using accurate mass measurements of the disulfide-paired ions and ECD-generated peptides with commercially available instrumentation (LTQFT and Nanomate). The data acquisition was easy to automate and allowed for hands off screening of a large number of HPLC fractions. Experimental conditions were developed to maximize the cleavage of the disulfide bonds while minimizing peptide backbone cleavage, which gives very distinctive ECD spectra from the disulfide-linked peptides. Because the ECD fragment ion masses will add up to the parent ion mass taking into account the addition of a hydrogen, this made it a simple task to scan through and find cysteine-paired peptides manually, especially for single paired peptides. Certainly a software-assisted approach similar to matching peptide identification could be employed to automate the process totally. Using just this approach, we were able to assign four disulfide pairs from the complex SUATM129 protein using a single HPLC run. Combined with secondary protease digestions and additional accurate mass spectrometry, we were able to assign nearly all (7 of 9) disulfide bonds as well as identify the carbohydrate-modified N-linked glycosylation sites of SUATM129 using these methods (Fig. 8).
It should be noted that in screening the HPLC fractions, a number of alternate cysteine pairs could be detected from lower background level signals. Recognizing the possibility of disulfide rearrangement during protein purification and the protease digestion steps despite great effort to keep these bonds stable, we relied on the UV absorbance of the HPLC peaks to determine that we were analyzing the most abundant cysteine-paired components in the digests. In addition, we repeatedly detected the same dominant disulfide pairs in repeat experiments using two different proteases to digest the protein (data not shown). Because MS analysis does not necessarily correspond to peptide abundance, several cycles of Edman chemical sequencing were performed on peptides in the HPLC fractions to confirm that the expected disulfide-linked peptides were the dominant component of that fraction (data not shown). Finally, similar MS studies on the SUA fragment of SUA-rIgG (38) also identified the same disulfide-bonded peptide pairs found in the SUA glycoprotein region of this study (data not shown).
Using SUATM129 as a representative, biologically active, but secreted protein, we propose a model of the disulfide bond and the N-linked glycosylation patterns of the subgroup A ASLV envelope glycoprotein (Fig. 8). As predicted from published data, the SUA glycoprotein subunit is bound to the TM glycoprotein subunit by a covalent disulfide bond: we found the first cysteine residue in SUA, C1 (Cys25), bound to the last extracellular cysteine in TM, C19 (Cys438). Six intrasubunit disulfide bonds define the secondary structure of the SUA glycoprotein, organizing the hr1, hr2, and vr3 variable domains for the critical interactions with their respective receptor to initiate entry. A disulfide bond was definitively assigned between Cys60 (C3) and Cys91 (C4), Cys131 (C5) and C6 (Cys184), C7 (Cys197) and C8 (Cys206), and C13 (Cys292) and C14 (Cys309). Disulfide bonds could not be definitively assigned to the adjacent cysteines in SUA, C11 (Cys258) and C12 (Cys259). The data show that a peptide containing C9 (Cys221) was bound to a peptide that contained C10 (Cys247), C11 and C12 and that further protein digestion proved that C9 was not bound to C10. Thus, there is a disulfide bond between C9 and C11 or C12, and a bond between C10 and the remaining C11/C12 residue. Two intrasubunit disulfide bonds were assigned in the TM glycoprotein: between C15 (Cys348) and C16 (Cys385), and C17 (Cys430) and C18 (Cys437). This proposed disulfide bond pattern leaves C2 (Cys38) as a free cysteine.
In this study, we have characterized a secreted form of the subgroup A ASLV envelope glycoprotein. Because the positions of the cysteine residues and the N-linked glycosylation sites are remarkably conserved between the subgroup A to E ASLV glycoproteins, we believe that the secondary structures of these glycoproteins will also be conserved including the position of the free cysteine C2, Cys38. The exact function of this free cysteine in ASLV fusion and entry is currently unknown. Smith and Cunningham found that binding of the subgroup A ASLV envelope glycoprotein with the Tva receptor induced the formation of a cysteine thiolate (Cys-S-) at the Cys38 position (28). They found that chemical inactivation of the Cys-S- or genetic substitution of Cys38 completely blocked ASLV fusion and infection. However, they determined that the Cys-S- does not mediate isomerization of the SUA and TM disulfide bond, nor is it required for the interaction of TM with a target membrane or the formation of helical bundles at low pH. Thus, some other step of the fusion process is dependent on the receptor-dependent formation of the cysteine thiolate.
In an earlier study, we mapped the modified N-linked glycosylation sites of the SUA glycoprotein fused to a rabbit IgG domain (SUA-rIgG) produced in DF-1 cells and purified from the cell culture supernatant using mass spectrometry (38). Ten of the 11 N-linked glycosylation sites were modified in SUA-rIgG, with only N8 (Asp236) unmodified. The SUATM129 protein contains both the SUA glycoprotein, and the extracellular region of the TM glycoprotein, had 11 of the 13 possible N-linked glycosylation sites modified by carbohydrate as detected by the conversion of a modified asparagine to aspartic acid after PNGase F digestion. The same 10 sites identified as modified in the extracellular SU glycoprotein in this study were also modified in a similar biologically active protein SUA-rIgG: only N8 (Asp236) was not modified with carbohydrate. One of the two possible N-linked glycosylation sites in the TM glycoprotein was modified: N12 (Asp392). Asp440 (N13) was not modified with carbohydrate possibly due to its close proximity to the CX6CC motif in the linker between the heptad repeats. Glycosylation of this site may inhibit the proposed flexibility of this region thought necessary for the conformational changes occurring during fusion.
It has been noted that although the amino acid sequences of the fusion glycoproteins of the Ebola virus GP2 and ASLV TM are very different, the organization of the secondary structures required for efficient fusion of the viral and cellular membranes are conserved (26). Both fusion proteins have cysteine residues flanking an internal fusion peptide region near the N terminus, followed by a region critical for conformational changes leading to fusion: two α-helical heptad repeat regions separated by a flexible linker region containing a CX6CC motif (39, 40). In addition, both the Ebola and ASLV envelope glycoproteins are trimers of heterodimers, GP1 and GP2 for Ebola, SU and TM for ASLV, with the heterodimers connected by a disulfide bond (41, 42). The disulfide bond pattern of the Ebola GP1/GP2 heterodimer has been determined biochemically with some of the bonds verified by crystal structure (41–43). Previously, based on the homology with Ebola GP2, the organization and disulfide bond pattern of ASLV TM were predicted to be the same as GP2. The data from this study have formally demonstrated that both Ebola GP2 and ASLV(A) TM glycoproteins have the same disulfide bond pattern organizing the functional domains in the fusion glycoproteins: a disulfide bond linking the cysteines flanking the internal fusion peptide, a disulfide bond between the first and second cysteine residues of the CX6CC motif, and the last cysteine in the CX6CC motif providing the covalent bond between the TM glycoprotein and the first cysteine of the surface glycoprotein Ebola GP1 and ASLV SUA.
The structural data presented here will be useful as a guide for future experiments designed to study the entry and fusion mechanism of ASLV envelope glycoproteins as a model for enveloped virus entry. The organization of the putative receptor binding domains (hr1, hr2, and vr3), the proof of the organization of the substructures in the TM fusion glycoprotein, and the identification of the disulfide bond linking the SU and TM heterodimers will aid in constructing new mutants to test function and to obtain atomic structure.
This work was supported, in whole or in part, by National Institutes of Health Grant AI48682. This work was also supported by the Mayo Clinic.
- ASLV
- avian sarcoma and leukosis virus
- CID
- collision-induced dissociation
- ECD
- electron capture dissociation
- ESI
- electrospray ionization
- FT
- Fourier transform
- FTICR
- Fourier transform ion cyclotron resonance
- MS/MS
- tandem mass spectrometry
- PNGase F
- N-glycosidase F
- SU
- surface
- TCEP
- tris (2-carboxyethyl) phosphine
- TEV
- tobacco etch virus
- TM
- transmembrane.
REFERENCES
- 1. Harrison S. C. (2008) Nat. Struct. Mol. Biol. 15, 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. White J. M., Delos S. E., Brecher M., Schornberg K. (2008) Crit. Rev. Biochem. Mol. Biol. 43, 189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hunter E. (1997) in Retroviruses (Coffin J. M., Hughes S. H., Varmus H. E. eds) pp. 71–120, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 4. Weiss R. A. (1992) in The Retroviruses (Levy J. A. ed) pp. 1–108, Plenum Publishing Corp., New York [Google Scholar]
- 5. Matsuyama S., Delos S. E., White J. M. (2004) J. Virol. 78, 8201–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mothes W., Boerger A. L., Narayan S., Cunningham J. M., Young J. A. T. (2000) Cell 103, 679–689 [DOI] [PubMed] [Google Scholar]
- 7. Smith J. G., Mothes W., Blacklow S. C., Cunningham J. M. (2004) J. Virol. 78, 1403–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Swanstrom R., Wills J. W. (1997) in Retroviruses (Coffin J. M., Hughes S. H., Varmus H. E. eds) pp. 263–334, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 9. Young J. A. T. (2001) in Fields Virology (Knipe D. M., Howley P. M. eds) pp. 87–103, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 10. Markosyan R. M., Bates P., Cohen F. S., Melikyan G. B. (2004) Biophys. J. 87, 3291–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Melikyan G. B., Barnard R. J., Abrahamyan L. G., Mothes W., Young J. A. T. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8728–8733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tamm L. K. (2003) Biochim. Biophys. Acta 1614, 14–23 [DOI] [PubMed] [Google Scholar]
- 13. Coffin J. M. (1992) Curr. Top. Microbiol. Immunol. 176, 143–164 [DOI] [PubMed] [Google Scholar]
- 14. Bates P., Young J. A., Varmus H. E. (1993) Cell 74, 1043–1051 [DOI] [PubMed] [Google Scholar]
- 15. Young J. A., Bates P., Varmus H. E. (1993) J. Virol. 67, 1811–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adkins H. B., Brojatsch J., Naughton J., Rolls M. M., Pesola J. M., Young J. A. T. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11617–11622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adkins H. B., Brojatsch J., Young J. A. T. (2000) J. Virol. 74, 3572–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brojatsch J., Naughton J., Rolls M. M., Zingler K., Young J. A. T. (1996) Cell 87, 845–855 [DOI] [PubMed] [Google Scholar]
- 19. Elleder D., Stepanets V., Melder D. C., Senigl F., Geryk J., Pajer P., Plachý J., Hejnar J., Svoboda J., Federspiel M. J. (2005) J. Virol. 79, 10408–10419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Damico R., Bates P. (2000) J. Virol. 74, 6469–6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hernandez L. D., Peters R. J., Delos S. E., Young J. A., Agard D. A., White J. M. (1997) J. Cell Biol. 139, 1455–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Holmen S. L., Salter D. W., Payne W. S., Dodgson J. B., Hughes S. H., Federspiel M. J. (1999) J. Virol. 73, 10051–10060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holmen S. L., Federspiel M. J. (2000) Virology 273, 364–373 [DOI] [PubMed] [Google Scholar]
- 24. Holmen S. L., Melder D. C., Federspiel M. J. (2001) J. Virol. 75, 726–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barnard R. J., Young J. A. T. (2003) Curr. Top. Microbiol. Immunol. 281, 107–136 [DOI] [PubMed] [Google Scholar]
- 26. Gallaher W. R. (1996) Cell 85, 477–478 [DOI] [PubMed] [Google Scholar]
- 27. Weissenhorn W., Calder L. J., Wharton S. A., Skehel J. J., Wiley D. C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6032–6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith J. G., Cunningham J. M. (2007) PLoS Pathog. 3, e198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Einfeld D. A., Hunter E. (1997) J. Virol. 71, 2383–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Himly M., Foster D. N., Bottoli I., Iacovoni J. S., Vogt P. K. (1998) Virology 248, 295–304 [DOI] [PubMed] [Google Scholar]
- 31. Schaefer-Klein J., Givol I., Barsov E. V., Whitcomb J. M., VanBrocklin M., Foster D. N., Federspiel M. J., Hughes S. H. (1998) Virology 248, 305–311 [DOI] [PubMed] [Google Scholar]
- 32. Federspiel M. J., Crittenden L. B., Hughes S. H. (1989) Virology 173, 167–177 [DOI] [PubMed] [Google Scholar]
- 33. Blum H., Beier H., Gross H. J. (1987) Electrophoresis 8, 93–99 [Google Scholar]
- 34. Ochsenbauer-Jambor C., Delos S. E., Accavitti M. A., White J. M., Hunter E. (2002) J. Virol. 76, 7518–7527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Mol. Cell Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 36. Zubarev R. A. (2004) Curr. Opin. Biotechnol. 15, 12–16 [DOI] [PubMed] [Google Scholar]
- 37. Zubarev R. A., Horn D. M., Fridriksson E. K., Kelleher N. L., Kruger N. A., Lewis M. A., Carpenter B. K., McLafferty F. W. (2000) Anal. Chem. 72, 563–573 [DOI] [PubMed] [Google Scholar]
- 38. Kvaratskhelia M., Clark P. K., Hess S., Melder D. C., Federspiel M. J., Hughes S. H. (2004) Virology 326, 171–181 [DOI] [PubMed] [Google Scholar]
- 39. Malashkevich V. N., Schneider B. J., McNally M. L., Milhollen M. A., Pang J. X., Kim P. S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 2662–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weissenhorn W., Carfí A., Lee K. H., Skehel J. J., Wiley D. C. (1998) Mol. Cell 2, 605–616 [DOI] [PubMed] [Google Scholar]
- 41. Jeffers S. A., Sanders D. A., Sanchez A. (2002) J. Virol. 76, 12463–12472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sanchez A., Yang Z. Y., Xu L., Nabel G. J., Crews T., Peters C. J. (1998) J. Virol. 72, 6442–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee J. E., Fusco M. L., Hessell A. J., Oswald W. B., Burton D. R., Saphire E. O. (2008) Nature 454, 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]