Figure 2.
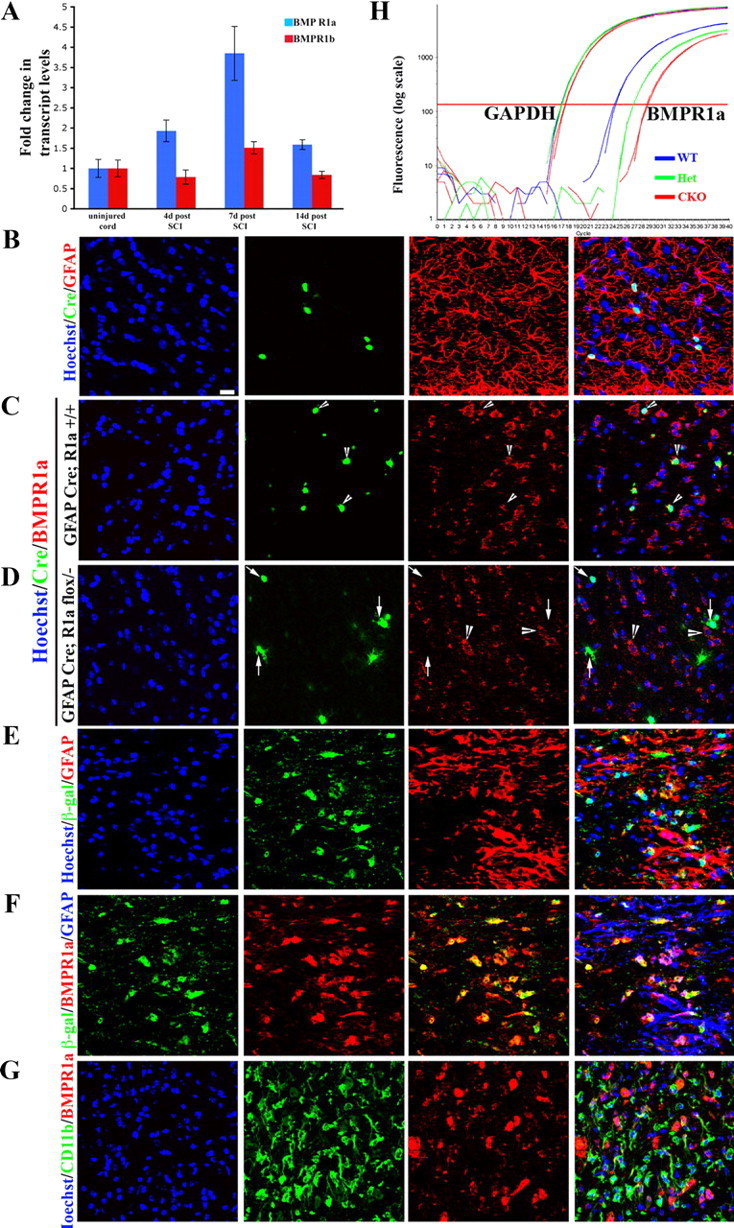
BMPR1a receptor is expressed on reactive astrocytes and not on inflammatory cells. A, Transcript levels of BMPR1a (blue bars) and BMPR1b (red bars) were normalized to GAPDH and expressed as fold changes compared with levels in uninjured controls± SEM. B, WT spinal cords stained with Hoechst nuclear stain (blue), Cre (green) and GFAP (red). Note that the Cre+ cells are GFAP+. C, D, Single confocal Z-sections from normal uninjured spinal cords stained with Hoechst nuclear stain (blue), Cre (green), and BMPR1a (red) from GFAP Cre (C) and GFAP Cre;BMPR1aflox/− (BMPR1a CKO) (D) animals. In GFAP Cre animals there is intact receptor staining seen on Cre+ cells (arrowheads). In the BMPR1a CKO animals, there is no staining for the receptor on Cre+ cells (arrows), while adjacent Cre-negative cells continue to express BMPR1a (D, arrowheads). E, Injured cord at 4 d post-SCI from a GFAP Cre, ROSA lacZ mouse stained with Hoechst nuclear stain (blue), β-galactosidase (green) and GFAP (red) showing that all the β-gal-positive cells are in GFAP-positive astrocytes. F, Sections in E stained with β-galactosidase (blue), BMPR1a (green), and GFAP (red). The β-gal staining clearly outlines the astrocytic soma, which stains for BMPR1a. G, Injured spinal cord sections at 4 d postinjury stained with Hoechst nuclear stain (blue), CD11b (green), and BMPR1a (red). Note that the receptor staining does not overlap with the CD11b. Scale bar (B–G), 20 μm. H, Real-time RT-PCR for exon2 of BMPR1a from total RNA extracted from spinal cords of WT, GFAP cre; BMPR1aflox/+ (het) and GFAP Cre; BMPR1aflox/− (CKO) mice. GAPDH serves as a loading control. There is an approximately 4-cycle reduction in the transcript levels of the floxed exon2.
