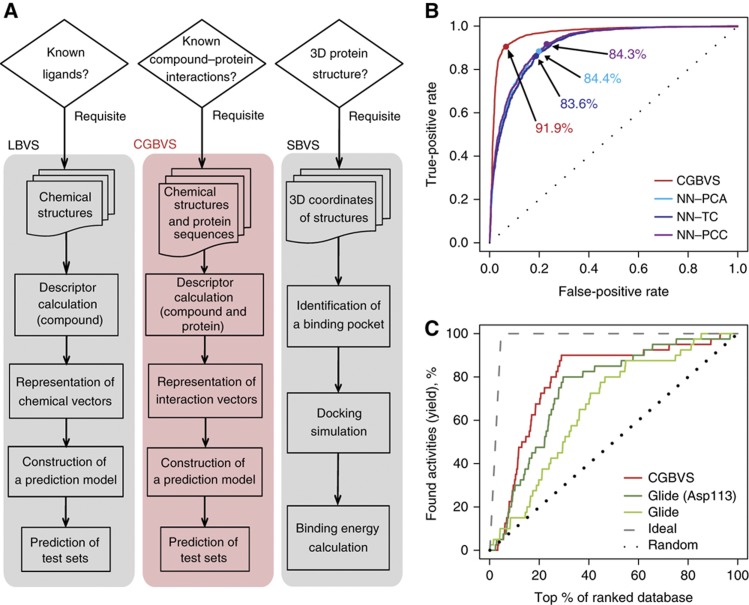
Figure 1.
Overview and performance of CGBVS. (A) Comparison of the strategies used for CGBVS, LBVS, and SBVS. CGBVS used multiple CPI data, represented the CPI in vector form, and used SVM for CPI pattern learning. (B) ROC curves obtained by fivefold cross-validation using compound–GPCR interactions for the CGBVS (red) and LBVS methods. The best accuracy rate for each method is also shown. NN–PCA (light blue), nearest neighbor (NN) method with the Pearson correlation coefficient in the constructed space using principal component analysis (PCA); NN–TC (navy), NN method with the Tanimoto coefficient (TC); and NN–PCC (purple), NN method with the Pearson correlation coefficient (PCC) in the original space. (C) Enrichment curves obtained by the CGBVS and SBVS methods. Glide was used both without constraints and with constrained hydrogen bonding between the compounds and Asp113, a residue known to be crucial for ligand binding to ADRB2 (Strader et al, 1987). Information regarding interactions between ADRB2 and its ligands was not used in test set for machine learning for CGBVS.