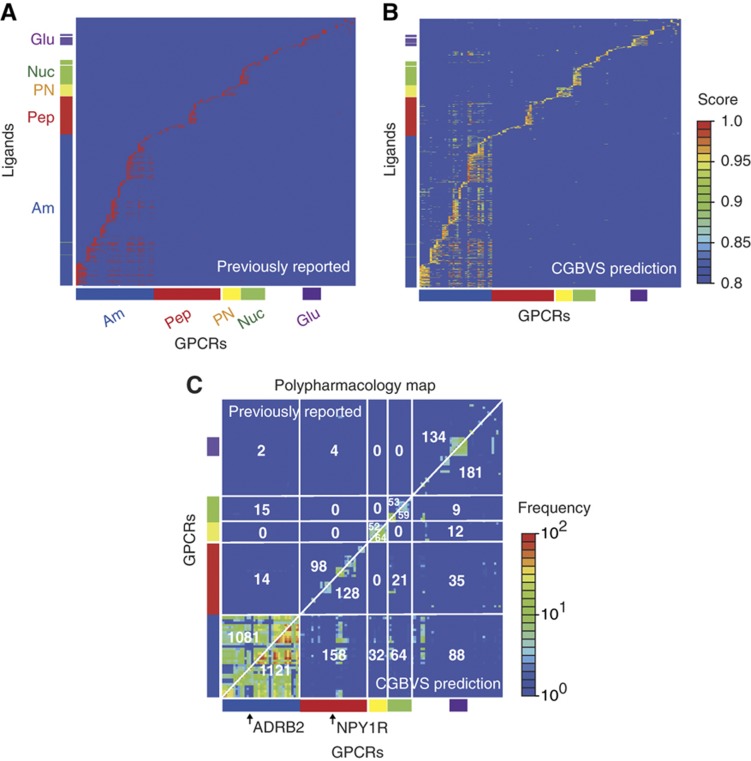
Figure 3.
Comparative polypharmacology maps for GPCRs. (A) Map of previously reported compound–GPCR interactions. Vertical and horizontal axes represent the compounds and the GPCRs, respectively. The reported CPIs are depicted as red dots. CGBVS used the CPIs as training data. The colored bars along each axis indicate the classes to which the compounds and GPCRs belong. Am, amines; Pep, peptides; PN, prostanoids; Nuc, nucleotides; and Glu, glutamates. (B) Map of predicted compound–GPCR interactions based on CGBVS. The CPIs are plotted with colors ranging from blue (low) to red (high), according to prediction scores. (C) Comparative polypharmacology map of GPCRs showing the number of shared compounds within a receptor family. The polypharmacology map was constructed as described by Paolini et al (2006) by plotting the numbers of common ligands for two given receptors. Previously reported and CGBVS-predicted interactions are shown in the upper-left and the lower-right diagonal halves, respectively. Each value indicates the number of common ligands for each GPCR subfamily. For example, 1081 compounds were reported to be ligands for amine receptors that cross-reacted with other amine receptors, and 14 amine receptor ligands were reported to cross-react with peptide receptors.