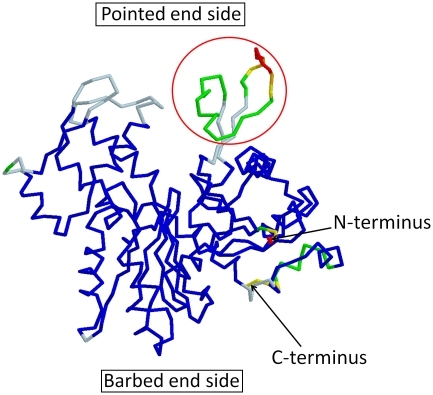
Figure 7.
The 88 crystal structures of monomeric actin were compared in an effort to identify flexible regions. The pdb IDs of these 88 structures are 1C0F, 1NWK, 1YAG, 2ASP, 2Q31, 2HF3, 1MA9, 2V52, 2VCP, 1D4X, 1P8Z, 1YVN, 2FXU, 2Q36, 2HF4, 1RFQ, 2VYP, 1DB0, 1QZ5, 1YXQ, 2GWJ, 2Q97, 3CHW, 1YXQ, 2FF3, 1DGA, 1QZ6, 1HLU, 2GWK, 2VCP, 3CI5, 1MDU, 3BUZ, 1EQY, 1RFQ, 1NLV, 2HMP, 2VYP, 3CJC, 2GWK, 2OAN, 1ESV, 1RGI, 1NM1, 2OAN, 2A3Z, 3DAW, 2HMP, 2OAN, 1H1V, 1S22, 1NMD, 2PAV, 2A40, 3EKS, 2OAN, 2V51, 1IJJ, 1SQK, 2A42, 2PBD, 2A41, 3EKU, 2Q1N, 2A40, 1J6Z, 1T44, 2A5X, 2Q0R, 2BTF, 3EL2, 2Q31, 1MDU, 1KXP, 1WUA, 2ASM, 2Q0U, 2D1K, 1LCU, 2V51, 1RDW, 1ATN, 1LCU, 1Y64, 2ASO, 2Q1N, 2FF6 and 1LOT. The structures were aligned against 1J6Z and the average and the s.d. of each α-carbon atom were calculated. Missing residues in the crystal structures were ignored in the calculation. The resultant averaged structure is presented. Residues with 0−0.5, 0.5−1, 1−2, 2−3, 3−4 and >4 Å s.d. were coloured in blue, light blue, green, yellow, orange and red, respectively. The residues located around the DNase I binding loop, indicated by a red circle, showed markedly large s.d. Furthermore, these residues were often missing in the crystal structures. In only 18 structures out of 88 structures, the main chain could be followed in this region, indicating that the area around the DNase I binding loop has significant flexibility.