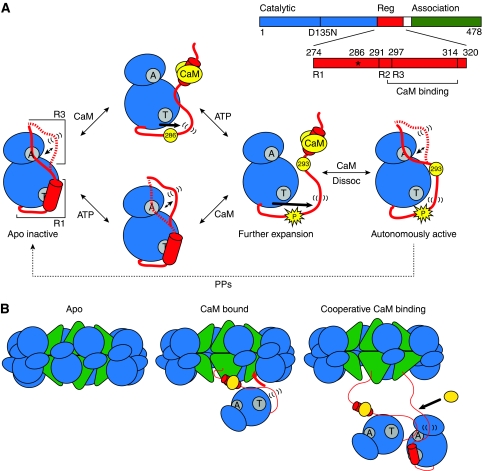
Figure 5.
(A) Model of the mechanism of autoinhibition and activation of CaMKII. The CaMKII monomer is represented with N- and C-lobes of the catalytic domain in blue and the regulatory domain in red. The ATP-binding and Thr286-docking sites are indicated in grey with letters ‘A’ and ‘T’, respectively. In the basal, apo intermediate, the R3 segment of the regulatory domain is in a dynamic equilibrium between docked and undocked, flexible (indicated by parentheses) conformations. ATP binding shifts the equilibrium towards the undocked conformation facilitating exposure of the catalytic cleft. CaM (yellow) binding releases autoinhibition and primes the regulatory domain for phosphorylation at residue 286. Autophosphorylation at Thr286 causes further flexibility of the regulatory domain and exposes site 293, which is implicated in CaM trapping. While CaM dissociation allows for reinstatement of the R3 equilibrium, the R1 helix remains predominantly unstructured. (B) Cartoon model of cooperative activation in the holoenzyme. CaM binding to one subunit causes undocking and unfolding of R1. The released R1 can displace the R3 segment of a neighbouring kinase subunit from the catalytic cleft. This R3 is undocked and primed for CaM association.