Abstract
Clustered regularly interspaced short palindromic repeat (CRISPR) is a recently discovered adaptive prokaryotic immune system that provides acquired immunity against foreign nucleic acids by utilizing small guide crRNAs (CRISPR RNAs) to interfere with invading viruses and plasmids. In Escherichia coli, Cas3 is essential for crRNA-guided interference with virus proliferation. Cas3 contains N-terminal HD phosphohydrolase and C-terminal Superfamily 2 (SF2) helicase domains. Here, we provide the first report of the cloning, expression, purification and in vitro functional analysis of the Cas3 protein of the Streptococcus thermophilus CRISPR4 (Ecoli subtype) system. Cas3 possesses a single-stranded DNA (ssDNA)-stimulated ATPase activity, which is coupled to unwinding of DNA/DNA and RNA/DNA duplexes. Cas3 also shows ATP-independent nuclease activity located in the HD domain with a preference for ssDNA substrates. To dissect the contribution of individual domains, Cas3 separation-of-function mutants (ATPase+/nuclease− and ATPase−/nuclease+) were obtained by site-directed mutagenesis. We propose that the Cas3 ATPase/helicase domain acts as a motor protein, which assists delivery of the nuclease activity to Cascade–crRNA complex targeting foreign DNA.
Keywords: Cas3, CRISPR, Ecoli subtype, site-directed mutagenesis, Streptococcus thermophilus
Introduction
Viruses have a major influence on all types of cellular life including eukaryotes, bacteria and archaea. To protect themselves against infection, prokaryotes have developed multiple defence barriers of various complexity, including prevention of adsorption, blocking of injection or degradation of the foreign nucleic acid (Sturino and Klaenhammer, 2006; Labrie et al, 2010). Recently, an adaptive microbial immune system, named clustered regularly interspaced short palindromic repeats (CRISPRs), has been identified that provides acquired immunity against viruses and plasmids (Barrangou et al, 2007). It consists of an array of short conserved DNA-repeat sequences that are interspaced by stretches of variable sequence called spacers, which generally originate from phage or plasmid DNA (Bolotin et al, 2005; Mojica et al, 2005). A set of cas (CRISPR-associated) genes is typically located in the vicinity to repeat-spacer array (Jansen et al, 2002; Makarova et al, 2006). CRISPR, in combination with the associated Cas proteins, forms the CRISPR/Cas systems, which are currently grouped into eight subtypes on the basis of clustering of cas genes (Haft et al, 2005; Makarova et al, 2006).
It was shown in Streptococcus thermophilus that during natural generation of phage-resistant variants, bacteria commonly alter their CRISPR loci by polarized incorporation of novel spacers, which originate from the phages used in the challenge (Barrangou et al, 2007; Deveau et al, 2008; Horvath et al, 2008). Mechanistically, the function of the CRISPR system has been dissected into two distinct steps: (i) CRISPR adaptation (immunization), which includes the recognition of foreign DNA followed by its subsequent processing and integration into the host chromosomal CRISPR locus and (ii) interference (immunity), which includes transcription of the CRISPR locus and generation of crRNAs that serve as the guide sequences in the binding and/or degradation of the target nucleic acid (Sorek et al, 2008; Hale et al, 2009; van der Oost et al, 2009; Horvath and Barrangou, 2010; Karginov and Hannon, 2010).
The molecular mechanism by which CRISPR provides resistance against foreign genetic elements is yet to be characterized. The first mechanistic details on the interference (immunity) step (Brouns et al, 2008) were obtained in experimental studies of the CRISPR/Cas system in Escherichia coli. E. coli K-12 strain possesses a single cluster of cas genes (Ecoli subtype) encoding eight proteins: three core proteins, Cas1, Cas2 and Cas3, and a subtype-specific five-protein set, Cse1–Cse2–Cse4–Cas5e–Cse3. The latter proteins are also referred to as CasABCDE and constitute a so-called Cascade complex. It was demonstrated (Brouns et al, 2008) in E. coli K-12 strain that the CRISPR region is transcribed into a long transcript (termed pre-crRNA), which is processed by the Cascade complex into small crRNAs, which contain a single spacer flanked on both sides by short nucleotide stretches originating from the repeat. These crRNA bound to the Cascade complex serve as the guide sequences that target foreign DNA and result in the CRISPR interference, presumably through the binding and/or degradation of the target nucleic acid.
In E. coli, the Cascade complex alone is able to generate crRNA but requires Cas3 to achieve immunity against foreign DNA (Brouns et al, 2008). Hence, Cas3 appears to be a key protein of CRISPR system, which is necessary for crRNA-guided interference of virus proliferation. Moreover, cas3 is present in most CRISPR/Cas subtypes (except Nmeni and Mtube) (Haft et al, 2005; Makarova et al, 2006) but the mechanism by which Cas3 achieves its function is not yet known. In silico analysis predicts that cas3 encodes a polypeptide comprised of HD-type phosphohydrolase/nuclease and DExD/H-box helicase domains (Haft et al, 2005; Makarova et al, 2006); however, so far there is no experimental evidence to demonstrate its role in vitro. Detailed mechanistic and structural studies of Cas3 have been limited, in part, by the lack of a suitable model system for carrying out quantitative biochemical and structural studies in vitro. Due to its poor solubility and aggregation, the E. coli Cas3 protein can only be expressed in a very limited amount (Gasiunas and Siksnys, unpublished data). The HD-type nuclease subunit of the related Sulfolobus solfataricus Cas3 protein has recently been expressed in a soluble form as a fusion protein and demonstrated to catalyse hydrolysis of double-stranded (ds)DNA and RNA, although no experimental details are yet available on the activity of the putative helicase domain (Han and Krauss, 2009).
In this paper, we provide the first biochemical characterization of the Cas3 protein identified in the CRISPR4 system of S. thermophilus DGCC7710. The latter CRISPR system belongs to the Ecoli subtype and comprises eight genes arranged similarly to the CRISPR system of E. coli (Horvath and Barrangou, 2010) (Figure 1A). We have cloned and expressed the cas3 gene of S. thermophilus DGCC7710 in E. coli ER2267, and purified the Cas3 protein to an apparent homogeneity. Here, we provide the first direct experimental evidence that Cas3 is a multifunctional protein possessing single-stranded DNA (ssDNA)-stimulated ATPase, ATP-dependent helicase and a metal-dependent single-stranded nuclease activities.
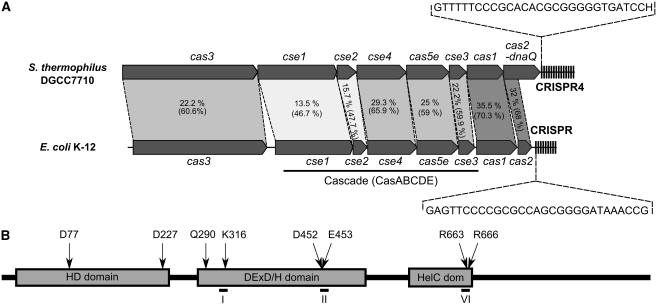
Figure 1.
(A) Schematic organization of the CRISPR4/Cas system of S. thermophilus DGCC7710 and the CRISPR/Cas system of E. coli K-12. Percentage of identical and similar (in parenthesis) residues between corresponding protein sequences that are connected by dashed lines. Conserved repeat sequences of each system are shown in the inserts. (B) Domain architecture of the S. thermophilus Cas3 protein. Domains identified by in silico analysis are shown as grey boxes. HD domain denotes HD-type phosphohydrolase/nuclease domain; DExD/H domain denotes DExD/H-box helicase domain; HelC dom denotes the C-terminal helicase domain. Conserved residues characteristic of the different domains and subject to alanine mutagenesis are indicated above the boxes. Location of the conserved helicase motifs are indicated by numbers I, II and VI.
Results
Expression and purification of Cas3 protein
According to the genomic sequence of S. thermophilus DGCC7710, the cas3 gene of the CRISPR4/Cas system encodes a protein of 926 amino acids with a predicted molecular mass of ∼106 kDa. The sequence data have been submitted to the GenBank database under accession No. HQ453272. In silico analysis of Cas3 protein reveals a multidomain architecture (Figure 1B). The N-terminal part of Cas3 shows conserved residues characteristic of the HD family of metal-dependent phosphohydrolases (HD domain) (Aravind and Koonin, 1998), whereas the C-terminal part of Cas3 has motifs characteristic of the Superfamily 2 (SF2) helicases (Singleton et al, 2007). A BLAST search revealed that the Cas3 protein from S. thermophilus DGCC7710 shows ∼22% identity and ∼60% similarity at the amino-acid level to the Cas3 protein of E. coli K-12 (Figure 1).
The cas3 gene from S. thermophilus DGCC7710 was cloned into the pBAD24-C(His)6 plasmid to yield a construct encoding a fusion protein containing a C-terminal 6-His-tag. The recombinant plasmid was expressed in the E. coli strain ER2267 and the Cas3 protein purified from the crude cell extracts. The purified Cas3 protein was ∼95% homogeneous as evaluated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) and Coomassie Blue staining (Supplementary Figure S2). Typically, ∼12 mg of protein was obtained in a single run from 1 l of E. coli culture.
Cas3 shows an ssDNA-stimulated ATPase activity
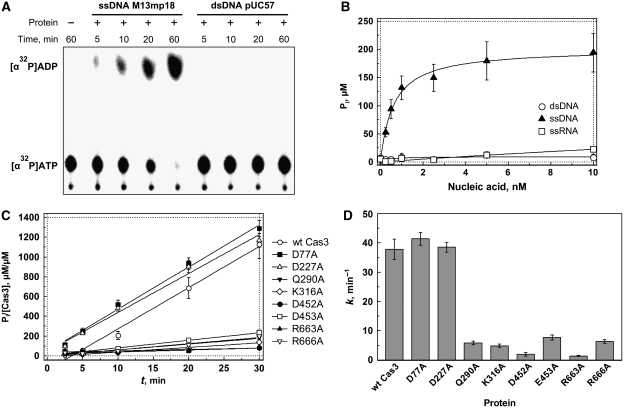
The presence of the characteristic helicase motifs (I, II and VI) responsible for ATP binding and hydrolysis in the primary sequence of Cas3 predicts that the protein would be an ATPase. The ability of Cas3 to hydrolyze ATP was first examined by monitoring the hydrolysis of radioactively labelled [α32P]ATP (Figure 2A). In the presence of the Cas3 protein alone, only minimal ATPase activity was detected. However, the ATP hydrolysis rate increased significantly in the presence of M13 circular ssDNA. We further investigated the ATP hydrolysis by Cas3 in the presence of ssDNA, dsDNA and RNA by measuring the concentration of accumulated phosphate product using a colorimetric assay (Hyun et al, 2008). Data analysis revealed that ATPase activity of Cas3 increased significantly in the presence of ssDNA but was not stimulated by the dsDNA or by RNA (Figure 2B). The linear accumulation of inorganic phosphate as a function of time (representative trace shown in Figure 2C) allowed us to calculate the rate of ATP hydrolysis at 0.5 mM of ATP as ∼38 min−1. Taken together, these results indicate that Cas3 possesses an ssDNA-dependent ATPase activity.
Figure 2.
Cas3 ATPase activity. (A) Radioactive ATPase assay. ATPase reactions were conducted at 30°C in a reaction buffer containing 10 mM Tris–HCl (pH 7.5 at 25°C), 30 mM KCl, 5% (v/v) glycerol, 2 mM MgCl2, 0.1 mg/ml BSA, 0.5 mM ATP, 2.5 nM ssDNA (M13mp18) or dsDNA (supercoiled form of pUC57 plasmid), and 250 nM Cas3. Reaction mixtures were supplemented with [α32P]ATP (5 Ci/mmol), spotted onto a polyethyleneimine-cellulose thin-layer plate and separated by chromatography followed by phosphorimager visualization. (B) ATP hydrolysis dependence of nucleic acids. Malachite green assay was used to measure ATP hydrolysis through the detection of liberated-free phosphate from ATP. Reaction mixtures in the buffer described above contained varying amounts of ssDNA (M13mp18), dsDNA (supercoiled form of pUC57 plasmid) or 2223 nt RNA. (C) Time courses of ATP hydrolysis. Reaction mixtures contained 3 nM ssDNA (M13mp18). Malachite green assay was used to measure ATP hydrolysis through the detection of liberated-free phosphate from ATP. (D) ATP hydrolysis rates. Reaction rate constant k (min−1) calculated from slopes of times courses shown in (C).
To test whether the conserved residues of the helicase domain are important for the Cas3 ATPase activity, we generated a set of alanine-substitution mutants. We mutated amino-acid residues Q290A (located in Q-motif (Tanner et al, 2003) important for ATP binding), K316A (located in motif I involved in ATP binding), D452A and E453A (located in motif II involved in Mg2+ coordination at the ATPase active site), R663A and R666A (located in motif VI (Tanner and Linder, 2001) involved in ATP binding). As illustrated in Figure 2D, the ATPase activity of the mutants decreased significantly, indicating the importance of the conserved amino-acid residues for the ATP binding/hydrolysis. Conversely, mutations in the N-terminal HD domain had no effect on the ATPase activity of Cas3.
In order to test the nucleotide specificity of Cas3, we compared the catalytic activity using ribo- and deoxyribonucleotide cofactors (Supplementary Figure S3). Cas3 exhibited a strong preference towards ATP or dATP. We also found that GTP and dGTP could be hydrolyzed in the presence of ssDNA, but with specific activities that were approximately 10-fold lower than observed with ATP. No significant hydrolytic activity was observed with other nucleotides. Study of the divalent metal ion dependence of the Cas3 ATPase activity revealed that the fastest rate was observed with Mn2+, followed closely by Mg2+ or Ca2+. Cu2+ supported hydrolysis at a much lower rate. No significant ATPase activity above background was observed using Co2+, Zn2+ or Ni2+ (Supplementary Figure S4).
Cas3 shows nuclease activity located in the HD domain
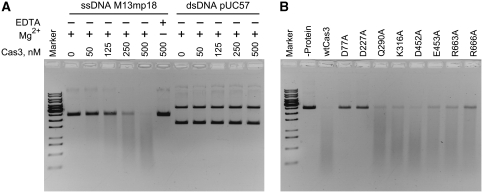
The nuclease activity of Cas3 was analysed using circular M13mp18 ssDNA or pUC57 supercoiled double-stranded plasmid DNA (Figure 3). Cas3 degraded the M13mp18 ssDNA in a concentration- and time-dependent manner (Figure 3; Supplementary Figure S5). In contrast, virtually no hydrolysis of the dsDNA occurred during the 2-h incubation. Mg2+ ions but not ATP were required for the ssDNA hydrolysis. Indeed, ssDNA was degraded with similar rates both in the absence or in the presence of ATP (data not shown).
Figure 3.
Cas3 nuclease activity. (A) Degradation of the ssDNA and dsDNA. Various amounts of Cas3 were incubated in the presence of 4 nM of M13 ssDNA or pUC57 dsDNA at 37°C for 2 h in the presence (+) or absence (−) of 10 mM MgCl2 or 10 mM EDTA. (B) Effect of mutations on Cas3 nuclease activity. In all, 500 nM of protein was incubated in the presence of 4 nM of M13 ssDNA at 37°C for 2 h.
Conserved amino-acid residues H27, H76, D77 and D276 located in the N-terminal HD-like domain (Aravind and Koonin, 1998) of Cas3 (Figure 1B; Supplementary Figure S1) were identified as being part of the putative active site responsible for divalent metal binding and ssDNA hydrolysis. Alanine replacement mutants D77A and D227A were constructed by site-directed mutagenesis, mutant proteins were purified and their ability to degrade DNA was analysed (Figure 3B). Experimental data indicate that while ssDNA was fully degraded in 2 h by the wild type (wt) Cas3, ssDNA hydrolysis was significantly reduced for D77A and D227A. Mutations in the helicase domain had much weaker effects on the associated nuclease activity (Figure 3B).
Cas3 shows DNA unwinding activity
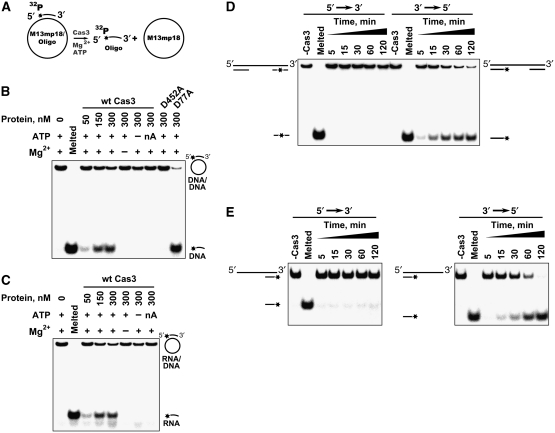
An ssDNA-dependent ATPase activity of Cas3 suggests a possible translocase/helicase function. To analyse whether the purified Cas3 protein possesses true helicase activity (i.e. DNA strand separation), we determined its capacity to unwind duplex DNA substrates (Figure 4). Since many helicases require a stretch of ssDNA in order to load onto the substrate and since Cas3 ATPase activity is stimulated by ssDNA, the substrate was constructed by hybridizing a 20-nt oligodeoxynucleotide, labelled with 32P at its 5′-terminus, to a circular M13mp18 ssDNA that contained a complementary sequence (Matson, 1986) (Figure 4A). This substrate was incubated with Cas3 and the reaction products were separated on a polyacrylamide gel. The labelled oligonucleotide was then detected by autoradiography. The experimental results indicate (Figure 4B) that Cas3 possesses a DNA unwinding activity that is dependent upon the presence of both Mg2+ and ATP but is not supported in the presence of the non-hydrolysable ATP analogue 5′-adenylyl-β, γ-imidodiphosphate (AMP-PNP). Furthermore, the Cas3 nuclease-deficient mutant D77A unwound the DNA similarly to the wt protein while the D452A mutant, which has compromised ATPase activity, showed no unwinding activity.
Figure 4.
Cas3 helicase activity and polarity. (A). Schematic representation of duplex unwinding assay. (B, C) DNA–DNA and RNA–DNA duplex unwinding by Cas3. Cas3 displacement of a P32-labelled 20 nt oligodeoxynucleotide (B) or 22 oligoribonucleotide (C) annealed to an ssM13mp18 DNA is monitored in the polyacrylamide gel. Reactions were performed at 30°C for 1 h in the reaction buffer: 10 mM Tris–HCl (pH 7.5 at 25°C), 25 mM KCl, 15% (v/v) glycerol, 1 mM MgCl2, 0.1 mg/ml BSA, 2 mM ATP, 0.5 nM substrate and various amounts of protein. nA denotes the ATP analogue AMP-PNP. D452A and D77A are ATPase and nuclease domain mutants, respectively. (D) Cas3 polarity assay I. Cas3 displacement of 30 nt double-stranded fragments at the ends of the linear M13mp18 DNA. Partial duplex DNA was prepared by EheI cleavage of the labelled 60 nt oligodeoxynucleotide annealed to the M13pm18 DNA. Duplex regions are separated by a single-stranded region of few thousand nucleotides. Reaction mixture contained 500 nM of Cas3. Reactions were stopped at defined time intervals. (E) Cas3 polarity assay II. Cas3 displacement of the oligonucleotide-based 73 nt substrates containing 53 nt 3′- or 5′-overhangs. Reactions were performed as in Figure 4B and C except that with the oligonucleotide-based substrates 500 nM of Cas3, 1.5 nM of substrate and 250 nM trap DNA (unlabelled oligonucleotide) were used in the reaction.
To test whether the Cas3 protein unwinds an RNA/DNA heteroduplex, a substrate was constructed by hybridizing a 22-nt oligoribonucleotide to M13mp18 ssDNA as mentioned above. The experimental results indicate (Figure 4C) that Cas3 displaces the 22-nt oligoribonucleotide in the presence of ATP and Mg2+ ions. Thus, Cas3 can be classified as both a DNA–DNA and DNA–RNA helicase.
With partially duplex substrates, each DNA helicase is thought to bind first to an ssDNA region and then to approach and unwind duplex DNA in a particular direction. To determine the directionality of Cas3, we first prepared a pair of labelled helicase substrates (Figure 4D) that contained duplex DNA at both ends of a long linear ssDNA molecule. Since the substrates comprise duplex regions at both ends of a long linear molecule, Cas3 must first bind to the internal single-stranded regions of these substrates. If the enzyme subsequently moves into a 3′–5′ direction along the ssDNA segment, it would displace the 5′-labelled fragment from the substrate. In contrast, the 3′-labelled fragment would be displaced if the enzyme migrates in a 5′–3′ direction. Experimental data presented in Figure 4 indicate that Cas3 moves primarily 3′ to 5′ along the ssDNA segment.
We also performed an alternative unwinding assay using radiolabelled DNA substrates of different structures. These 73 nt partial oligoduplexes contained 53 nt 3′- or 5′-single-stranded overhangs in addition to a 20-bp duplex region (Figure 4E). Cas3 could only unwind the substrates containing a 3′-overhang, confirming the 3′–5′ polarity seen above. The unwinding activity of Cas3 was observed in the presence of ATP and was not detected in the absence of ATP or in the presence of the non-hydrolyzable ATP analogue (AMP-PNP), suggesting that ATP hydrolysis is required for helicase function.
Discussion
cas3 is a core gene present in most of the CRISPR/Cas subtypes (except Nmeni and Mtube). The presence of the HD-type phosphohydrolase/nuclease domain led to the proposal that Cas3 might catalyse crRNA-guided cleavage of foreign nucleic acids (van der Oost et al, 2009). Since Cas3 also carries an apparent ATP/helicase domain, it was suggested (Marraffini and Sontheimer, 2010) that it could act also as a target DNA unwindase to enable protospacer hybridization with the crRNA. Here, we overproduced the Cas3 protein of a S. thermophilus CRISPR4/Cas system (Horvath and Barrangou, 2010), which belongs to the Ecoli subtype, purified Cas3 to ∼95% homogeneity, and performed functional characterization in vitro.
Cas3 degrades ssDNA
The Cas3 protein of S. thermophilus is arranged as a polypeptide comprised of an N-terminal HD-type phosphohydrolase/nuclease domain and a C-terminal SF2 helicase domain (Haft et al, 2005; Makarova et al, 2006). This architecture is characteristic of most Cas3 proteins but it is not absolutely conserved. For example, in the Cas3 protein of Apern subtype, the domains are found in swapped order, while in the Cas3 of Ypest, the HD-type phosphohydrolase/nuclease domain is found at the N-terminus of the cas2 gene (Brouns et al, 2008). In some cases, such as in S. solfataricus, the HD-type phosphohydrolase/nuclease and the DExD/H-box helicase domains are encoded as separate genes (Han et al, 2009).
The HD-type phosphohydrolase/nuclease domain is found in a superfamily of enzymes with either a predicted or known phosphohydrolase activity (Aravind and Koonin, 1998). According to Prosite database, bacterial HD domains are found in combination of 49 different partner domains, which presumably modulate protein function. HD-domain-containing proteins appear to be involved in nucleic acid metabolism and signal transduction, as well as other unknown functions (Aravind and Koonin, 1998). For example, the HD domain of the E. coli tRNA nucleotidyltransferase exhibits 2,3-cyclic phosphodiesterase, 2-nucleotidase and phosphatase activities (Yakunin et al, 2004).
The N-terminal domain of Cas3 contains a characteristic signature of the HD-type phosphohydrolase/nuclease domain (Figure 1B). The conserved residues (H…HD…D) predicted to be involved in the coordination of the divalent metal (Aravind and Koonin, 1998) are conserved in the N-terminal domain of S. thermophilus Cas3 and correspond to the residues H27, H76, D77 and D227. We show here that in the presence of magnesium ions, Cas3 has a nuclease activity that degrades ssDNA. It does not act on dsDNA (Figure 3). While Mg2+ ions are absolutely necessary for the nuclease activity, ATP is not. Mutation of the key metal-coordinating residues D77 and D227 compromised the ability of Cas3 to degrade ssDNA, but did not affect the ATPase activity, consistent with in silico predictions.
The ssDNA-degrading activity of the S. thermophilus Cas3 protein is in contrast to the apparent dsDNA and dsRNA cleavage activity of the standalone HD phosphohydrolase/nuclease of S. solfataricus (Han and Krauss, 2009) with only minor degradation of ssDNA or ssRNA observed. It is not clear whether this represents a true difference between the CRISPR systems or whether the activity of the HD domain is altered when the helicase domain is present (even though helicase activity is not required).
Cas3 unwinds DNA in the presence of ATP
The C-terminal fragment of Cas3 protein carries signature motifs characteristic of the SF2 helicases of the DExD/H subgroup (Singleton et al, 2007). Helicases use ATP to unwind and translocate nucleic acids or remodel nucleic acids or nucleic acid–protein complexes (Cordin et al, 2006; Singleton et al, 2007; Fairman-Williams et al, 2010). We show here that the Cas3 protein exhibits ssDNA-dependent ATPase activity, which absolutely requires Mg2+ ions. Nine conserved domains Q, I, Ia, Ib and II–VI have been identified in the SF2 group of DExD/H-type helicases (Cordin et al, 2006; Singleton et al, 2007; Fairman-Williams et al, 2010). The highest level of sequence conservation in the SF1 and SF2 helicase families is seen in the residues that coordinate binding and hydrolysis of the triphosphate (motifs I, II and VI). Mutations of the amino-acid residues Q290, K316, D452, E453, R663 and R666 in the conserved motifs I, II and VI, abolished or significantly compromised ATP hydrolysis. However, none of the helicase mutants showed any change in the ssDNA-degrading activity. Among different deoxy- and ribonucleotides tested, only ATP or dATP were hydrolysed significantly (Supplementary Figure S3). The maximum rate of ATP hydrolysis was ∼38 molecules per minute (Figure 2D). This value is also close to the value reported for the bacterial XPB helicase (Biswas et al, 2009) and NS3 helicase from hepatitis C virus (Kyono et al, 2003). The inefficient ATP hydrolysis suggests that Cas3 alone is not very processive or that it is an intrinsically slow ATPase involved, for example, in the local remodelling of protein–crRNA complex (see below).
In parallel to the ATPase activity, Cas3 protein is able to unwind oligonucleotide–M13 DNA complex. The unwinding activity of Cas3 is dependent on the protein concentration and presence of ATP and Mg2+ ions. Using a linear M13 molecule with a large internal region of ssDNA where a helicase can assemble, and two 32P-labelled duplex regions of different lengths, we show that Cas3 helicase has 3′ → 5′ directionality. It is likely that Cas3 functions as DNA translocase but translocation activity still has to be demonstrated directly. Cas3 contains a long C-terminal extension that follows the conserved helicase domains of the SF2 superfamily. Terminal domains of helicases have been demonstrated to direct recruitment of partner proteins/complexes, to promote interactions with other proteins, or to facilitate recognition of specific nucleic acid regions (Karow and Klostermeier, 2010). One cannot exclude that the C-terminal domain of Cas3 performs similar functions.
Putative role of Cas3 in the CRISPR mechanism
Taken together, our biochemical studies show that Cas3 protein combines in a single protein both ssDNA nuclease and ATPase/helicase activities. How can these activities be reconciled with the CRISPR mechanism? It was previously shown that in E. coli at least, a so-called Cascade complex consisting of the five protein set Cse1–Cse2–Cse4–Cas5e–Cse3 performs processing of a pre-crRNA into mature crRNAs, which remain associated with Cascade (Brouns et al, 2008). Hence, Cas3 is not required in the crRNA generation step and is probably involved in the CRISPR adaptation and/or interference steps. Indeed, in vivo studies demonstrate that both Cas3 and Cascade–crRNA complexes are required for the interference step (Brouns et al, 2008).
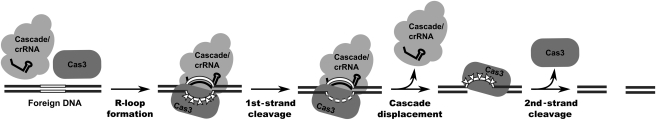
We suggest that ATP-dependent DNA helicase activity of Cas3 can contribute both to the protospacer sequence recognition by the Cascade–crRNA complex and DNA cleavage (Figure 5). It is possible that Cascade–crRNA complex interactions with the target DNA promote strand separation and create ssDNA regions for the Cas3 binding, which further unwinds DNA strands promoting crRNA hybridization to the complementary protospacer DNA. Indeed, Cas3 ATPase activity, which is necessary for the strand displacement, is stimulated by ssDNA but not by dsDNA (Figure 2). Invasion of the crRNA into the DNA duplex should result in the formation of the R-loop structure (Camps and Loeb, 2005) and the ATP-dependent Cas3 helicase activity may promote the formation of such loops. On the other hand, in the R-loop structure, a single-stranded antisense DNA strand may become a target for the nuclease domain of Cas3. Indeed, we show that Cas3 degrades closed circular ssDNA but shows nearly no activity on the dsDNA. The nuclease activity of the Cas3 resides in the N-terminal HD domain. Thus, tandem action of the Cascade–crRNA and Cas3 protein on the R-loop structure may result in the single-strand break in the protospacer region. One may speculate that after cleavage of one DNA strand, Cas3 helicase activity further remodels the Cascade–crRNA complex, for example, by displacing the Cascade–crRNA, so creating a platform for the second DNA strand cleavage by the HD domain to generate a double-strand break (Garneau et al, 2010).
Figure 5.
Proposed Cas3 mechanism of action. Assembly of the ternary Cascade–crRNA–Cas3 complex with the target DNA promotes strand separation and crRNA hybridization to the complementary DNA strand to generate the R-loop structure. A single-DNA strand in the R-loop structure is cut at multiple sites by Cas3 nuclease resulting in the single-strand break in the protospacer region. After cleavage of one DNA strand, Cas3 helicase activity may further remodel the Cascade–crRNA complex, for example, by displacing the Cascade–crRNA, and create a platform for the second DNA strand cleavage by the HD domain to generate a double-strand break.
Interestingly, despite the fact that Cascade recognizes protospacers with and without a protospacer adjacent motif (PAM) (Mojica et al, 2009) equally well, only a protospacer flanked by a PAM is subject to interference. This suggests that interference is not just due to binding of Cascade to the target DNA—blocking transcription or replication—but to subsequent interference events. One cannot exclude the possibility that the Cas3 C-terminal domain may contribute to PAM recognition. Cas3 nuclease/helicase studies using substrates mimicking the crRNA–DNA complexes are a subject of future research in our laboratory.
Materials and methods
Cloning, expression and purification of Cas3
S. thermophilus DGCC7710 genomic DNA was used as a template in PCR reactions to clone cas3 into the pBAD24-CHis expression vector (kindly provided by R Sukackaite) via NcoI and XhoI sites to generate a pCas3 plasmid, which was used for the overexpression of the C-terminal (His)6-tagged Cas3 protein variant. The following primers were used for the PCR amplification of cas3: 5′-AGGTCTCACATGAAACATATTAATGATTATTTTTGGGC-3′ and 5′-ACTCGAGAACCGACTCATTCTTATCCAAC-3′. Full sequencing of cas3 gene in pCas3 plasmid revealed no difference with the original cas3 sequence. Cas3 was expressed in E. coli ER2267 strain grown in LB broth supplemented with ampicillin (100 μg/ml) and kanamycin (25 μg/ml). Cells were grown at 37°C to OD600 of ∼0.5, the growth temperature was decreased to 16°C and expression induced with 0.2% (w/v) arabinose for 20 h. Harvested cells were disrupted by sonication and cell debris were removed by centrifugation. The supernatant was loaded onto the Ni2+-charged 5 ml HiTrap chelating HP column (GE Healthcare) and eluted with a linear gradient of increasing imidazole. The fractions containing Cas3 were pooled and subsequently loaded onto heparin column eluting using linear gradient of increasing NaCl concentration. The fractions containing Cas3 were pooled and dialysed against 10 mM Tris–HCl (pH 7.5), 300 mM KCl, 1 mM EDTA, 1 mM DTT, 50% (v/v) glycerol and stored at −20°C. The homogeneity of protein preparations was estimated by SDS–PAGE and western blotting against (His)6-tag with anti-His antibodies (Novagen). Concentrations of Cas3 and its mutants were determined by measuring absorbance at 280 nm using an extinction coefficient of 132 700/M/cm (Gill and von Hippel, 1989).
Mutagenesis
Conserved amino acids of HD hydrolase and DExD/H domains were identified by multiple alignment of Cas3 homologues using the COBALT tool (Papadopoulos and Agarwala, 2007). The mutants D77A, D227A, Q290A, K316A, D452A, E453A, R663A and R66A were obtained by the site-directed mutagenesis as previously described (Tamulaitis et al, 2007). Sequencing of the entire gene for each mutant confirmed that only the designed mutation had been introduced.
ATPase assay
ATPase reactions were conducted at 30°C in a reaction buffer containing 10 mM Tris–HCl (pH 7.5 at 25°C), 30 mM KCl, 5% (v/v) glycerol, 2 mM MgCl2, 0.1 mg/ml BSA, 0.5 mM ATP and 3 nM ssDNA (M13mp18) or dsDNA (supercoiled form of pUC57 plasmid) and 250 nM of Cas3 or mutant proteins. In the radioactivity assay, reaction mixtures were supplemented with [α32P]ATP (5 Ci/mmol) (Hartmann Analytic, Braunschweig, Germany). An aliquot (1 μl) was spotted onto a polyethyleneimine-cellulose thin-layer plate and ATP and resulting ADP were separated by chromatography in a 0.325-M KH2PO4 (pH 3.5) and visualized using a FLA-5100 phosphorimager (Fujilm, Tokyo, Japan).
Malachite green assays (BioAssay Systems) were used to measure ATP hydrolysis through the detection of liberated-free phosphate. Reaction mixtures in the buffer described above contained varying amounts of ssDNA (M13mp18 obtained from Fermentas, Vilnius, Lithuania), dsDNA (supercoiled form of pUC57 plasmid obtained from Fermentas) or 2223 nt RNA (obtained by transcription of the control template using a ‘T7 high yield transcription kit’, Fermentas) and 250 nM of Cas3 or its mutants. Reactions were initiated by adding enzyme to a mixture of the other reaction components. Aliquots were removed at fixed time intervals and the reaction was stopped by adding EDTA to a final concentration of 27 mM. The relationship between the absorbance and phosphate concentration was established by using KH2PO4 as a standard and used to calculate phosphate concentrations in the ATPase assays. ATPase rates are quoted as the mean of three independent experiments.
Helicase assay
Partial DNA or RNA–DNA duplexes were used in the helicase assay. Oligodeoxynucleotide 20-mer 5′-CCTGCAGGTCGACTCTAGAG-3′ and oligoribonucleotide 22-mer 5′-CAUGCCUGCAGGUCGACUCUAG-3′ were purchased from Metabion (Martinsried, Germany). The 5′-ends of oligonucleotides were radiolabelled using T4 polynucleotide kinase (PNK) (Fermentas) and [γ32P]ATP (Hartmann Analytic). Partial duplexes were assembled by mixing M13mp18 ssDNA and labelled complementary oligonucleotide at 1.5:1 molar ratio followed by annealing in 10 mM Tris–HCl (pH 7.5) buffer. The helicase reactions were performed at 30°C for 60 min in a buffer containing 10 mM Tris–HCl (pH 7.5), 25 mM KCl, 15% (v/v) glycerol, 1 mM MgCl2, 2 mM ATP (or AMP-PNP), 0.5 nM of substrates and indicated amounts of Cas3 or its mutants. Reactions were initiated by addition of Cas3. The reactions were terminated by addition of 3 × ‘stop’ solution (67.5 mM EDTA, 27% (v/v) glycerol, 0.3% (w/v) SDS, 0.03% (w/v) bromophenol blue). The products were separated by electrophoresis through 8% (w/v) non-denaturing polyacrylamide gel, and visualized using a FLA-5100 phosphorimager (Fujilm).
Helicase polarity assay
Two types of substrates were used in the Cas3 polarity assay. The M13mp18-based substrate was constructed by phosphorylating the 5′-ends of 5′-GCGCGGGGAGAGGCGGTTTGCGTATTGGGCGCCAGGGTGGTTTTT-3′ and 5′-CTTTTCACCAGCGAG-3′ oligonucleotides with [γ32P]ATP or ATP, respectively (or vice versa), followed by annealing to the complementary sequences of M13mp18 and ligation by T4 ligase (Fermentas). The resulting 60 bp partial duplexes were cut with EheI (Fermentas) to produce linear ssDNA molecules with short duplex regions at both ends.
For oligonucleotide-based helicase substrates, oligonucleotide 20-mers 5′-GGGGGGGGGGTAGTTGAGAA-3′ or 5′-CCCGCGCGCGTCGTCATGCG-3′ were labelled at the 5-end using PNK (Fermentas) and [γ32P]ATP, and annealed to the complementary sequence at the 5′- or 3′-end of the 73-mer 5′-GGGCGCGCGCAGCAGTACGCTAGTACTGTTCCCATGTCTAAGGAGGGGTTGCGTTCTCAACTACCCCCCCCCC-3′ to generate partial oligoduplexes containing 53 nt 3′ or 5′-overhangs.
Nuclease assay
Single-stranded M13mp18 DNA and double-stranded supercoiled form of pUC57 plasmid were used as substrates in the nuclease assay. Nucleic acid cleavage reactions were performed at 37°C for 120 min in buffer containing 10 mM Tris–HCl (pH 7.5), 60 mM KCl, 10% (v/v) glycerol, 10 mM MgCl2, 4 nM ssDNA of M13mp18 or dsDNA supercoiled form of pUC57 plasmid, and 500 nM Cas3 or mutant proteins. Reactions were initiated by addition of protein. The reactions were stopped by addition of phenol–chloroform followed by chloroform extraction. Aqueous fraction was mixed with 3 × loading solution (67.5 mM EDTA, 27% (v/v) glycerol, 0.3% (w/v) SDS). The products were separated by electrophoresis through 0.8% (w/v) agarose gels and visualized by the ethidium bromide staining.
Supplementary Material
Acknowledgments
We acknowledge Dr Mark Dillingham for providing the PcrA helicase sample, Dr Mark Szczelkun, Dr Mindaugas Zaremba and Dr Giedrius Sasnauskas for helpful discussions and comments on the manuscript. The work was supported by the Programme Gilibert between the Lithuanian Ministry of Education and Science and by French Ministry of Foreign and European affairs (MAEE) and French Ministry of Higher education and Research (MESR).
Footnotes
The authors declare that they have no conflict of interest.
References
- Aravind L, Koonin EV (1998) The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci 23: 469–472 [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 [DOI] [PubMed] [Google Scholar]
- Biswas T, Pero JM, Joseph CG, Tsodikov OV (2009) DNA-dependent ATPase activity of bacterial XPB helicases. Biochemistry 48: 2839–2848 [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD (2005) Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151 (Pt 8): 2551–2561 [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps M, Loeb LA (2005) Critical role of R-loops in processing replication blocks. Front Biosci 10: 689–698 [DOI] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P (2006) The DEAD-box protein family of RNA helicases. Gene 367: 17–37 [DOI] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S (2008) Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190: 1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, Jankowsky E (2010) SF1 and SF2 helicases: family matters. Curr Opin Struct Biol 20: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis M, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468: 67–71 [DOI] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182: 319–326 [DOI] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE (2005) A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP (2009) RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Krauss G (2009) Characterization of the endonuclease SSO2001 from Sulfolobus solfataricus P2. FEBS Lett 583: 771–776 [DOI] [PubMed] [Google Scholar]
- Han D, Lehmann K, Krauss G (2009) SSO1450—a CAS1 protein from Sulfolobus solfataricus P2 with high affinity for RNA and DNA. FEBS Lett 583: 1928–1932 [DOI] [PubMed] [Google Scholar]
- Horvath P, Barrangou R (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327: 167–170 [DOI] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R (2008) Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol 190: 1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun M, Bohr VA, Ahn B (2008) Biochemical characterization of the WRN-1 RecQ helicase of Caenorhabditis elegans. Biochemistry 47: 7583–7593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Embden JD, Gaastra W, Schouls LM (2002) Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43: 1565–1575 [DOI] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ (2010) The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell 37: 7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow AR, Klostermeier D (2010) A structural model for the DEAD box helicase YxiN in solution: localization of the RNA binding domain. J Mol Biol 402: 629–637 [DOI] [PubMed] [Google Scholar]
- Kyono K, Miyashiro M, Taguchi I (2003) Characterization of ATPase activity of a hepatitis C virus NS3 helicase domain, and analysis involving mercuric reagents. J Biochem 134: 505–511 [DOI] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE, Moineau S (2010) Bacteriophage resistance mechanisms. Nat Rev Microbiol 8: 317–327 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV (2006) A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ (2010) CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson SW (1986) Escherichia coli helicase II (urvD gene product) translocates unidirectionally in a 3′ to 5′ direction. J Biol Chem 261: 10169–10175 [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C (2009) Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155 (Pt 3): 733–740 [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E (2005) Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60: 174–182 [DOI] [PubMed] [Google Scholar]
- Papadopoulos JS, Agarwala R (2007) COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23: 1073–1079 [DOI] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB (2007) Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem 76: 23–50 [DOI] [PubMed] [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P (2008) CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol 6: 181–186 [DOI] [PubMed] [Google Scholar]
- Sturino JM, Klaenhammer TR (2006) Engineered bacteriophage-defence systems in bioprocessing. Nat Rev Microbiol 4: 395–404 [DOI] [PubMed] [Google Scholar]
- Tamulaitis G, Zaremba M, Szczepanowski RH, Bochtler M, Siksnys V (2007) Nucleotide flipping by restriction enzymes analyzed by 2-aminopurine steady-state fluorescence. Nucleic Acids Res 35: 4792–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner NK, Cordin O, Banroques J, Doere M, Linder P (2003) The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol Cell 11: 127–138 [DOI] [PubMed] [Google Scholar]
- Tanner NK, Linder P (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell 8: 251–262 [DOI] [PubMed] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ (2009) CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci 34: 401–407 [DOI] [PubMed] [Google Scholar]
- Yakunin AF, Proudfoot M, Kuznetsova E, Savchenko A, Brown G, Arrowsmith CH, Edwards AM (2004) The HD domain of the Escherichia coli tRNA nucleotidyltransferase has 2′,3′-cyclic phosphodiesterase, 2′-nucleotidase, and phosphatase activities. J Biol Chem 279: 36819–36827 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.