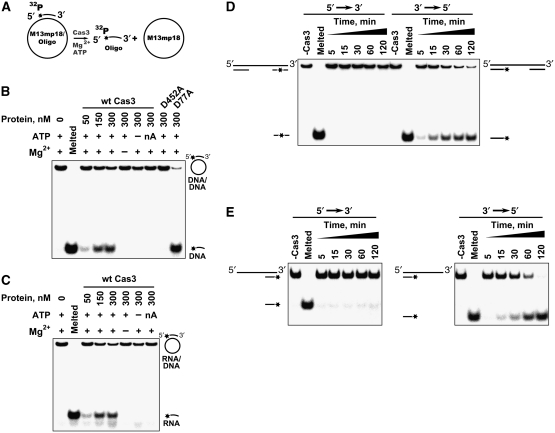
Figure 4.
Cas3 helicase activity and polarity. (A). Schematic representation of duplex unwinding assay. (B, C) DNA–DNA and RNA–DNA duplex unwinding by Cas3. Cas3 displacement of a P32-labelled 20 nt oligodeoxynucleotide (B) or 22 oligoribonucleotide (C) annealed to an ssM13mp18 DNA is monitored in the polyacrylamide gel. Reactions were performed at 30°C for 1 h in the reaction buffer: 10 mM Tris–HCl (pH 7.5 at 25°C), 25 mM KCl, 15% (v/v) glycerol, 1 mM MgCl2, 0.1 mg/ml BSA, 2 mM ATP, 0.5 nM substrate and various amounts of protein. nA denotes the ATP analogue AMP-PNP. D452A and D77A are ATPase and nuclease domain mutants, respectively. (D) Cas3 polarity assay I. Cas3 displacement of 30 nt double-stranded fragments at the ends of the linear M13mp18 DNA. Partial duplex DNA was prepared by EheI cleavage of the labelled 60 nt oligodeoxynucleotide annealed to the M13pm18 DNA. Duplex regions are separated by a single-stranded region of few thousand nucleotides. Reaction mixture contained 500 nM of Cas3. Reactions were stopped at defined time intervals. (E) Cas3 polarity assay II. Cas3 displacement of the oligonucleotide-based 73 nt substrates containing 53 nt 3′- or 5′-overhangs. Reactions were performed as in Figure 4B and C except that with the oligonucleotide-based substrates 500 nM of Cas3, 1.5 nM of substrate and 250 nM trap DNA (unlabelled oligonucleotide) were used in the reaction.