Table 1. Enrichment of the significant 16S eigenpositions in taxonomic groups.
| Eigenposition | Correlated | Anticorrelated | ||||||
| Taxonomic Group | j | J | P-value | Taxonomic Group | j | J | P-value | |
| 2 | Eukarya | 75 | 107 |
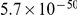
|
Bacteria | 75 | 175 |
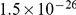
|
 Microsporidia Microsporidia |
||||||||
| 3 | Archaea | 57 | 57 |
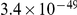
|
||||
 Microsporidia Microsporidia |
||||||||
| 4 | Gamma | 32 | 32 |
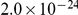
|
Actinobacteria | 72 | 74 |
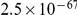
|
| Proteobacteria |
 Archaea Archaea |
|||||||
| 5 | Microsporidia | 29 | 36 |
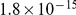
|
Archaea | 21 | 21 |
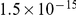
|
| 6 | Fungi/Metazoa | 32 | 32 |
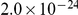
|
Rhodophyta | 26 | 26 |
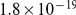
|
 Microsporidia Microsporidia |
||||||||
| 7 | Alveolata | 21 | 21 |
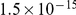
|
Fungi/Metazoa | 32 | 32 |
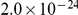
|
 Microsporidia Microsporidia |
||||||||
Probabilistic significance of the enrichment of the
k = 75 organisms, with largest relative
nucleotide frequency increase or decrease in each of the significant
eigenpositions, in the respective taxonomic groups. The
P-value of each enrichment is calculated as described [35], assuming
hypergeometric probability distribution of the J annotations
among the K = 339 organisms, and of the
subset of 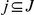 annotations among the subset of
k = 75 organisms.
annotations among the subset of
k = 75 organisms.
