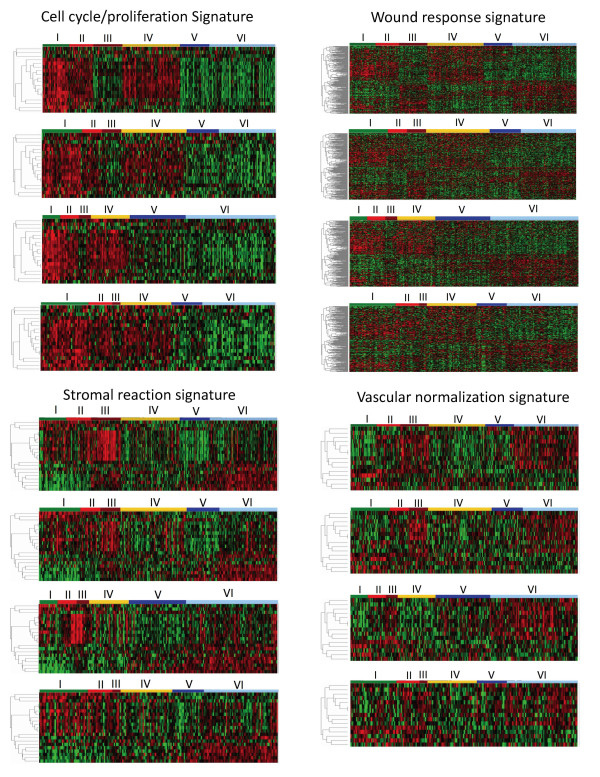
Figure 3.
Validation of molecular subtypes of breast cancer established in this study. One-way hierarchical clustering analysis was performed on 327 samples in our dataset using genes associated with cell cycle/proliferation, wound-response [9], stromal reaction [21], and tumor vascular endothelial normalization [22,23]. Breast cancer samples were arranged according to their subtype as shown at the top of each panel. Dendrograms of signature genes are shown on the left. The identities of genes in all four dendrograms are listed in the Additional file 3, Figure S4. None of the genes used in this study were part of the 783 probe-sets used for molecular subtyping. The same gene clusters generated from our dataset were used to draw heat maps for the other three independent datasets. The heat maps from top to bottom for each signature were KFSYSCC, EMC [10], Uppsala [19], and TRANSBIG [20]. Each molecular subtype shared the same distinctive gene expression pattern among all four datasets. Subtypes I, II and IV showed increased expressions of cell cycle/proliferation genes. Subtypes I and II showed higher expression of stromal genes known to associate with poorer survival [21]. Subtypes III and VI had elevated expression of genes associated with vascular endothelial normalization. The concordance of differential gene expression for the six molecular subtypes between the KFSYSCC dataset and each of the other three independent datasets [10,19,20] was analyzed by Pearson correlation. The p value for each correlation coefficient was determined by comparing with null distribution based on 10,000 permutations of each independent dataset at subtype level. The Pearson correlation coefficient between the KFSYSCC dataset and that of EMC, Uppsala or TRANSBIG was 0.94, 0.92 or 0.87 for cell cycle/proliferation, 0.85, 0.84 or 0.78 for wound response, 0.94, 0.91 or 0.87 for stromal reaction, and 0.86, 0.86 or 0.83 for tumor vascular endothelial normalization. All p values were < 0.0001.