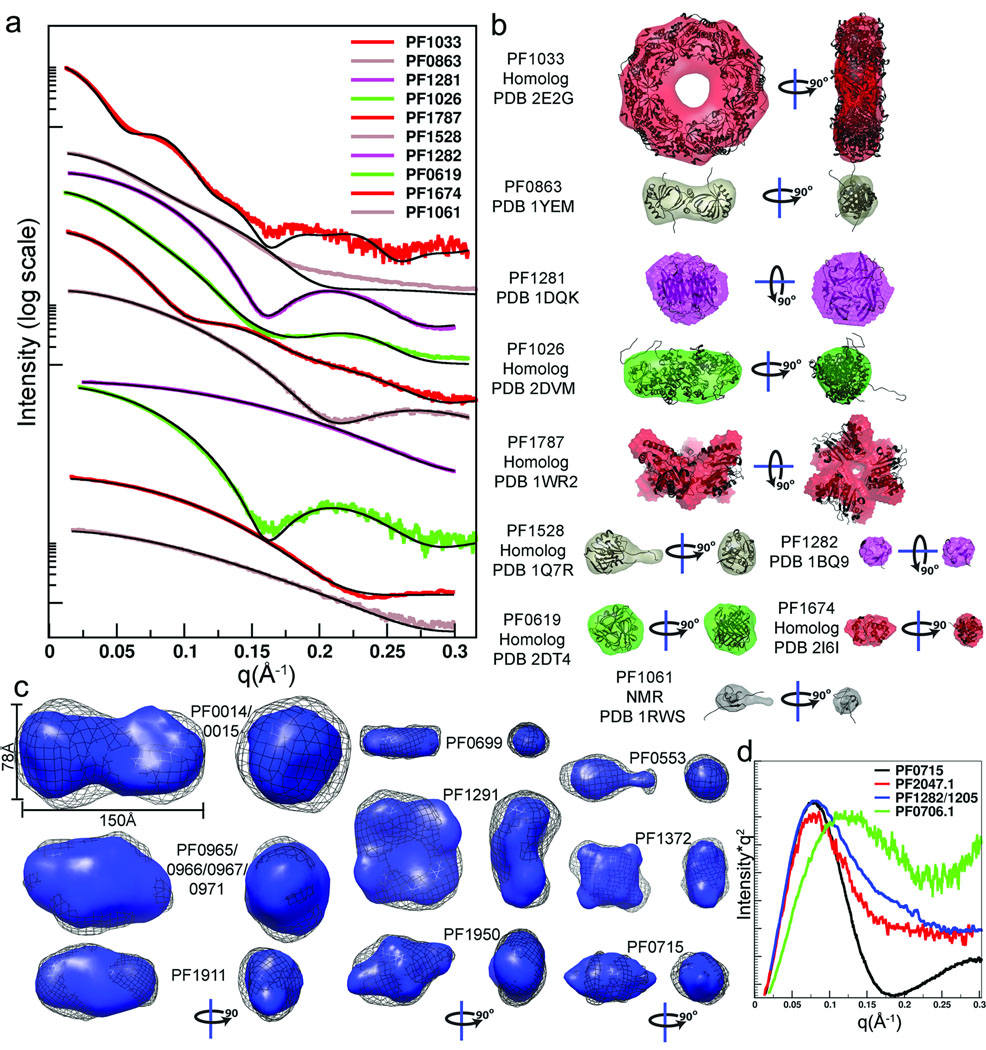
Figure 3.
SAXS provides accurate shape and assembly in solution for most samples. (a) For the ten proteins with structural homologs or existing structures, the experimental scattering data (colors) were compared with the scattering curve calculated for the matching structure (black). (b) For monodisperse samples, the envelope determinations (colored as in a) were overlaid with the existing structures (ribbons). All monomeric units had a seven amino-acid His-tag attached. (c) For the 9 proteins with no pre-existing structural information, envelope predictions from two independent programs were compared and generally agree. The DAMMIN results (black mesh) were generated without symmetry. The GASBOR results used 2-fold symmetry for PF0014/0015, PF0965/0966/0967/0971, PF1911 (dimer), PF00716 (dimer), PF0699 (dimer) and PF1950 (dimer). Four-fold symmetry was imposed on tetrameric PF1291 and PF1372. (d) Plotting the SAXS data as I*q2 vs. q (Kratky plot) highlights proteins with large unfolded regions. The Kratky plot of PF0715 is shown for comparison of a folded protein and shows characteristic parabolic behavior at wide angles. In contrast PF0706.1, PF2047.1, and PF1282/1205 have SAXS data consistent with unfolded regions as reflected in the non-parabolic wide-angle properties.