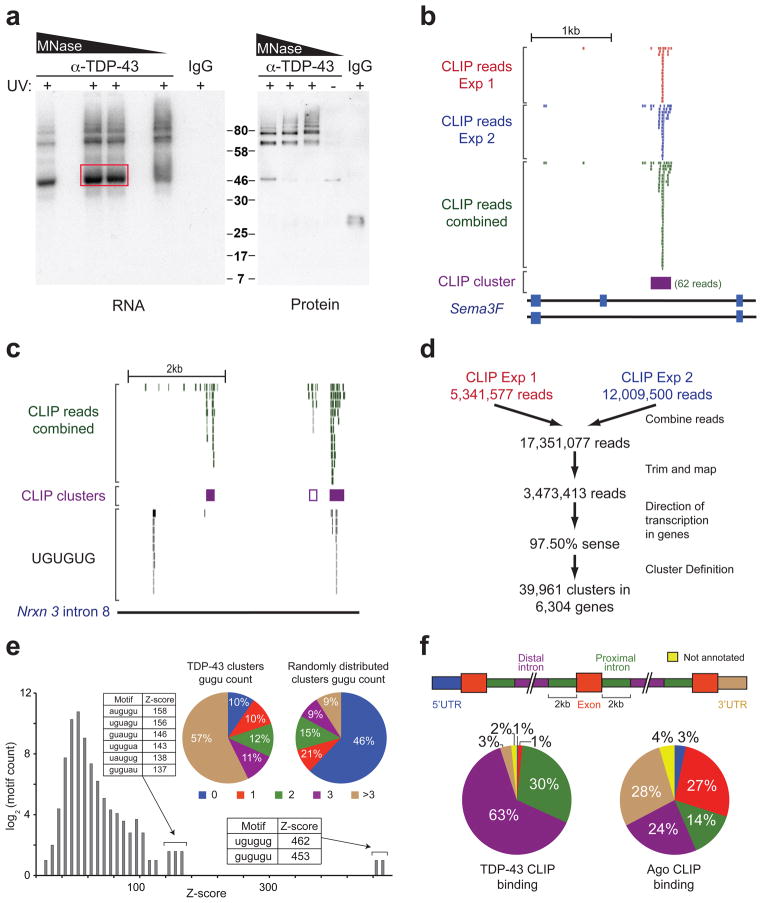
Figure 1. TDP-43 binds distal introns of pre-mRNA transcripts through UG-rich sites in vivo.
(a) Autoradiograph of TDP-43-RNA complexes trimmed by different concentrations of micrococcal nuclease (MNase) (left panel). Complexes within red box were used for library preparation and sequencing. Immunoblot showing TDP-43 in ~46kD and higher molecular weight complexes dependent on UV-crosslinking (UV) (right panel). (b) Example of a TDP-43 binding site (CLIP-cluster) on Semaphorin 3F defined by overlapping reads from 2 independent experiments surpassing a gene-specific threshold. (c) UCSC browser screenshot of neurexin 3 intron 8 (mm8; chr12:89842000-89847000), displaying four examples of TDP-43 binding modes. The right-most CLIP cluster represents a ‘canonical’ binding site coinciding GU-rich sequence motifs while the left-most cluster lacks GU-rich sequences and a region containing multiple GU-repeats shows no evidence of TDP-43 binding. The second CLIP cluster (middle purple-outlined box) with weak binding was found only when relaxing cluster-finding algorithm parameters. (d) Flow-chart illustrating the number of reads analyzed from both CLIP-seq experiments. (e) Histogram of Z-scores indicating the enrichment of GU-rich hexamers in CLIP-seq clusters compared to equally sized clusters, randomly distributed in the same pre-mRNAs. Sequences and Z-scores of the top 8 hexamers are indicated. Pie-charts enumerate clusters containing increasing counts of (GU)2 compared to randomly distributed clusters (p≈0, χ2=21,662). (f) Pre-mRNAs were divided into annotated regions (upper panel). Distribution of TDP-43 (left panel), or previously published Argonaute binding sites21 as a control (right panel), showed preferential binding of TDP-43 in distal introns.