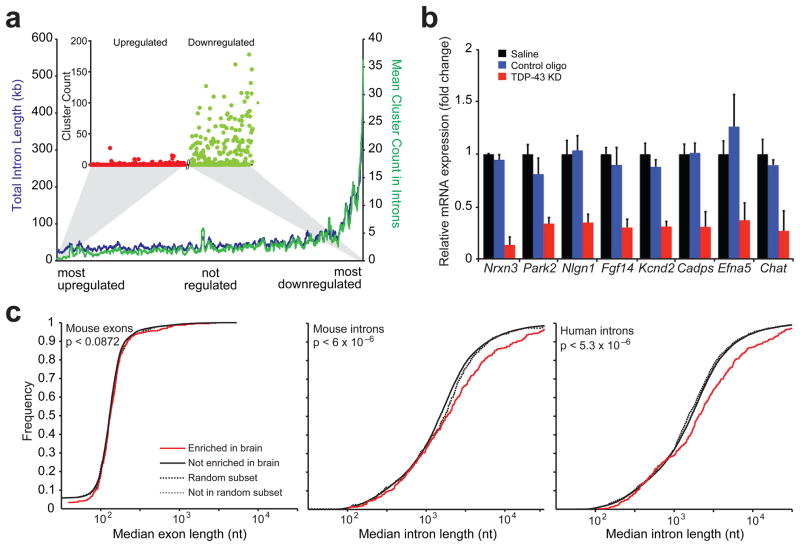
Figure 3. Binding of TDP-43 on long transcripts enriched in brain sustains their normal mRNA levels.
(a) Correlation between RNA-seq and CLIP-seq data. Genes were ranked upon their degree of regulation after TDP-43 depletion (x-axis) and the mean number of intronic CLIP clusters found in the next 100 genes from the ranked list were plotted (y-axis; green line). Similarly, the mean total intron length for the next 100 genes was plotted (y-axis; red line). The cluster count for each up-regulated gene (red dots) and each down-regulated gene (green dots) was plotted using the same ordering (inset). (b) Quantitative RT-PCR for selected down-regulated genes with long introns (except for Chat) revealed significant reduction of all transcripts when compared to controls (p < 8×10−3). Standard deviation was calculated within each group for 3–5 biological replicates. (c) Cumulative distribution plot comparing exon length (left panel) or intron length (middle panel) across mouse brain tissue enriched genes (388 genes) and non-brain tissue enriched genes (15,153 genes). Genes enriched in brain have significantly longer median intron length compared to genes not enriched in brain (solid red line and black lines, p-value < 6.2×10−6 by two-sample Kolmogorov Smirnov goodness-of-fit hypothesis test) while a random subset of 388 genes shows no difference in intron length (dashed lines). Similar analysis across human brain tissue enriched genes (387 genes) and non-brain tissue enriched genes (17,985 genes) also showed significantly longer introns in brain enriched genes (solid red and black lines, p < 5.3×10−6) while a random subset of 387 genes shows no difference in intron length (dashed lines) (right panel).