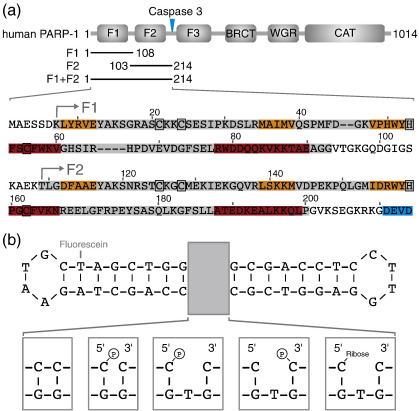
Fig. 1.
Protein and DNA constructs used in this study. (a) During apoptosis, PARP-1 is cleaved by caspase-322 into a 24-kDa fragment that contains the two N-terminal zinc fingers (F1 and F2) and an 89-kDa fragment composed of the third zinc finger (F3), the BRCT domain, the WGR domain and the C-terminal catalytic domain (the caspase-3 cleavage site is marked with a blue arrow). The expansion below shows the sequence of human PARP-1 residues 1–214, highlighting the fragments used in this study: F1 (residues 1–108), F2 (residues 103–214) and F1 + F2 (residues 1–214). Secondary structural elements are colored (α-helices, red; β-strands, orange), the caspase-3 recognition site is shown in blue and zinc-coordinating residues are indicated by black boxes. (b) Synthetic DNA dumbbell ligands used in this study. The same dumbbell scaffold was used to harbour different types of DNA strand breaks: a 5′-phosphorylated nick, a 5′-phosphorylated single nucleotide gap, a 3′-phosphorylated single nucleotide gap and a 5′-ribosylated single nucleotide gap (which results from strand incision of an abasic site during BER). Ligation of the nicked DNA dumbbell ligand produced a circular DNA without a strand break (left-handmost box).