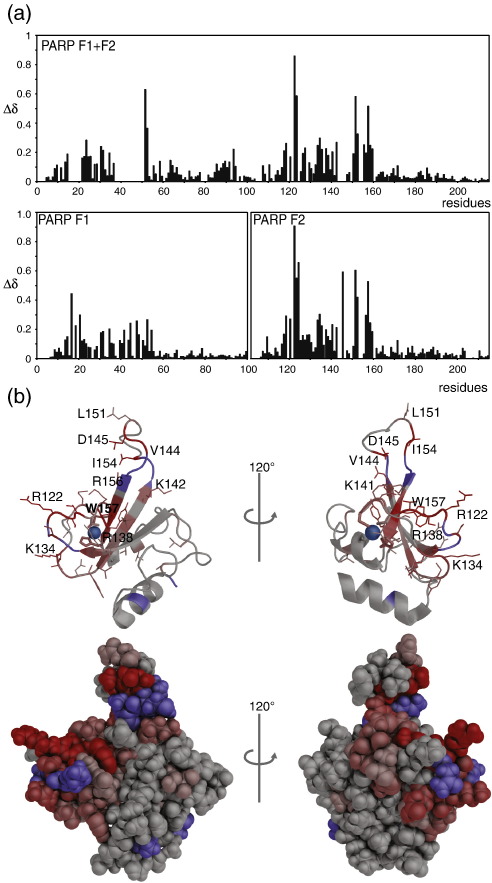
Fig. 8.
Chemical shift perturbation analysis. (a) Histograms of backbone amide chemical shift perturbations for F1 + F2 (upper panel), F1 and F2 (both lower panels) upon binding to the 45-nt 5′-phosphorylated gapped DNA dumbbell. Perturbations are calculated as Δδ = √[(δ1H)2 + (δ15N ÷ 5)2] (b) Chemical shift perturbations from (a) mapped onto the solution structure of F2 in both cartoon and space-filling representations. Perturbations were divided into five categories according to the SD over all perturbations: unaffected (0–0.5 × SD), weak (0.5–1 × SD), medium (1–1.5 × SD), strong (1.5–2 × SD) and very strong (> 2 × SD) and colored accordingly from gray (unaffected) to red (very strong). On the cartoon view, side chains of perturbed residues are drawn as thin lines. Residues for which no chemical shift perturbation could be determined due to missing assignments are colored blue.