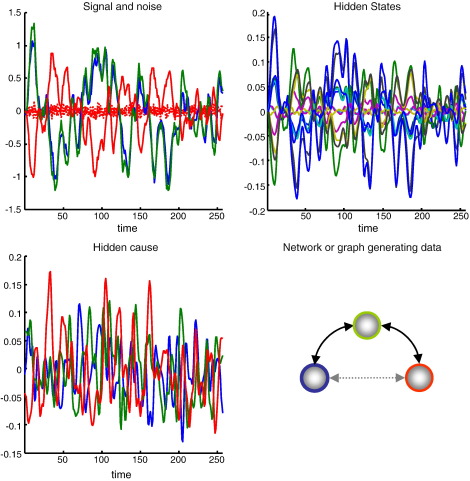
Fig. 2.
Synthetic data: This figure shows the synthetic data generated by a network or graph with three nodes. The upper left panel shows the simulated activity of each node over 256 (3.22 s) time bins. Signal is shown as solid lines and observation noise as dotted lines. The signal is a nonlinear function of the hidden hemodynamic and neuronal states shown on the upper right. In this model, there are five hidden states per node, which evolve according to the model's equations of motion. The dynamics seen here are caused by random neuronal fluctuations shown in the lower left panel. These were created by convolving a random Gaussian variable with a Gaussian convolution kernel of two bins standard deviation. The ensuing neuronal responses are communicated among nodes by extrinsic connections. In this example, we connected the blue node to the green node and the green node to the red node, as described in the main text. These influences or effective connectivity is denoted by the bidirectional solid black arrows. The light grey arrow denotes a possible but absent edge (anti-edge).