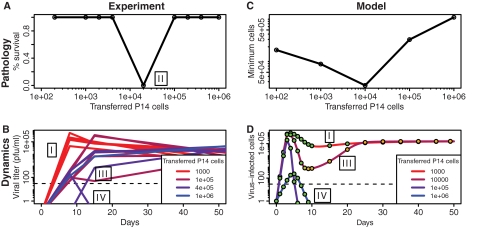
Fig. 4.
Pathology and dynamics of clone 13 LCMV infections. Experimental results of Blattman et al. (6) (A, B) can be qualitatively reproduced by our model (C, D). Mice were given adoptive transfers of different numbers of P14 cells (specific for the GP33 epitope) and subsequently infected with the clone 13 strain of LCMV, which typically gives rise to a persistent infection. Low initial numbers of P14 cells lead to chronic infection (outcome I). Intermediate initial numbers of P14 cells lead to the death of the mice around day 10 (outcome II). High numbers of transferred P14 cells lead to two possible outcomes: either the generation of an escape variant which forms a persistent infection (outcome III) or clearance of the virus (outcome IV). (D) In model time series, filled circles illustrate the relative amounts of wild-type to mutant virus, where green indicates that 100% of the cells were infected with the wild type and orange indicates that 100% were infected with the escape mutant. “Mutation” from a wild-type to an escape virus arises deterministically by starting at a low initial value (10−4) for a simulation using 104 transferred cells. We assume a 10% take of the transferred cells (5) in order to compare simulations to the experiment. Other parameters are the same as described for Fig. 1B except that α = 0.05/(target cells · day), β = 5 × 10−6/[(target cells)2 · day], k = 3.5 × 10−5 · day/(target cells · T cells · day), and s = 1.2/(T cells · day). Initial values were as follows: U = a/b, V1 = 1, V2 = 10−3 for simulation with 104 transferred cells (V2 = 0 otherwise), X1 = 10 × specified number of transferred cells, X2 = 102, and Q1 = Q2 = 0.