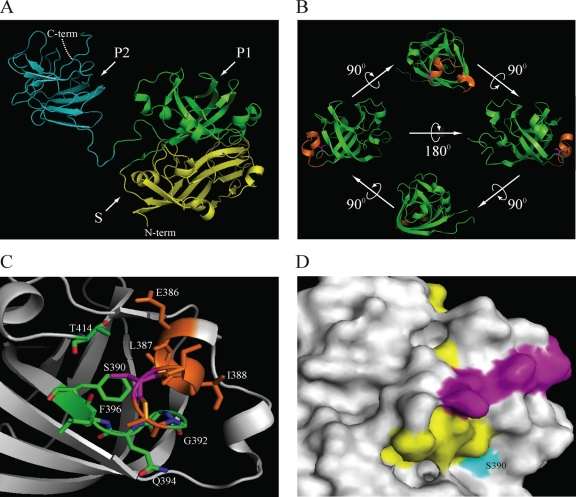
Fig. 7.
Structural location of S390 within the capsid protein of HEV. (A) Crystal structure of the truncated HEV capsid protein (residues 129 to 606) (PDB accession no. 2ZTN) (69). Domains are indicated as follow: shell (S) (residues 118 to 313) in yellow and subdomains P1 (residues 314 to 453) in green and P2 (residues 454 to 606) in cyan of the protruding (P) domain. (B) Ribbon diagram of the ternary P1 domain, showing the putative glycosylation site (376ADTLLGGLPTELISSA391, in orange). The S390 residue is shown in magenta and maps within a helix-turn-helix motif. (C) Schematic presenting the residues within a 4-Å distance from S390. Those residues that are part of the putative glycosylation site are in orange (386ELISSA391). Residues in green are part of the loop and β-sheet and comprise residues 392GGQLF396. The side chain of an additional residue, T414, located within a 4-Å distance is also indicated. (D) Surface map showing the location of residue S390 and linear epitopes of the 12A10 and 16D7 (purple) and 15B2 (yellow) monoclonal antibodies (23). Images were generated using PyMOL (7).