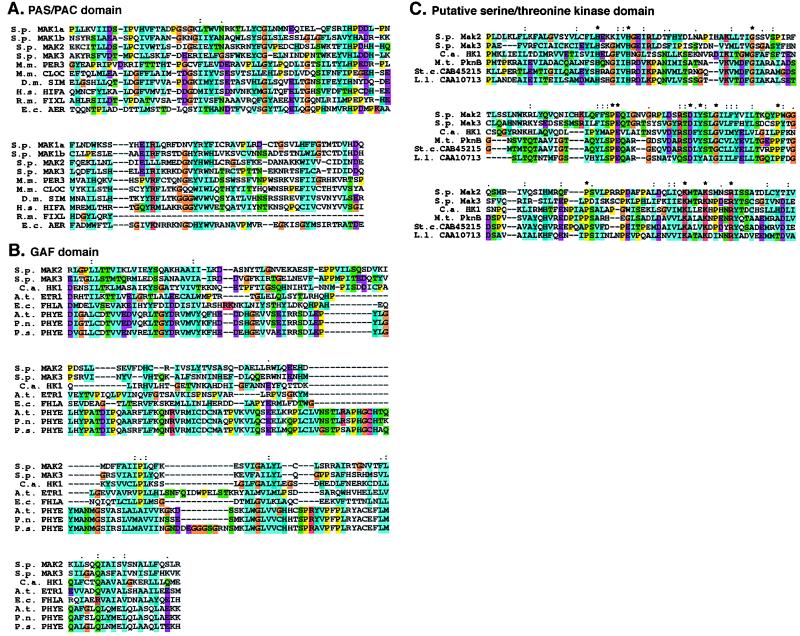
Figure 3.
Sequence alignment of Mak1p, Mak2p, and Mak3p. (A) Alignment of two PAS/PAC domains of S. pombe (S.p.) Mak1p (a and b) with similar domains in S. pombe (S.p.) Mak2p and Mak3p, Mus musculus (M.m.) PER3 and CLOCK, D. melanogaster (D.m.) SIM, human (H.s.) HIF1α, Rhizobium meliloti (R.m.) FixL, and E. coli (E.c.) Aer proteins. Homologies were identified by reiterative PSI-BLAST searches. Amino acid similarities are represented in color: yellow for proline, turquoise for hydrophobic residues (I, L, V, F, M, C), purple for acidic residues (E, D), red for basic residues (K, R), dark blue for aromatic residues (H, Y), green for polar residues (Q, S, T, N), and orange for glycine. Sequences are colored automatically on the basis of an alignment consensus that is calculated automatically by the CLUSTAL program. Residues that are completely conserved (*), highly conserved (:) or mostly conserved (.) are indicated. (B) Alignment of the GAF domain in S. pombe (S.p.) Mak2p and Mak3p with those in C. albicans (C.a.) HK1, Arabidopsis thaliana (A.t.) Etr1 and PhyE, E. coli (E.c.) FhlA, and Pharbitus nil (P.n.) and Pinus sylvestris (P.s.) PhyE proteins. Homologies are displayed as in A. (C) Atypical serine/threonine-kinase domains of S. pombe (S.p.) Mak2p and Mak3p, C. albicans (C.a.) HK1, M. tuberculosis (M.t.) PknB, and two uncharacterized proteins from Streptomyces coelicolor (St. c.; CAB45215) and Lactococcus lactis (L.l.; CAA10713). Homologies are displayed as in A.