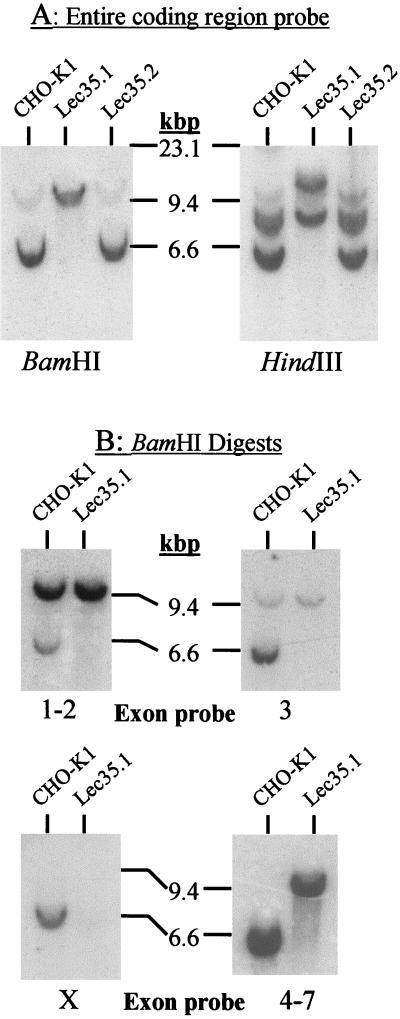
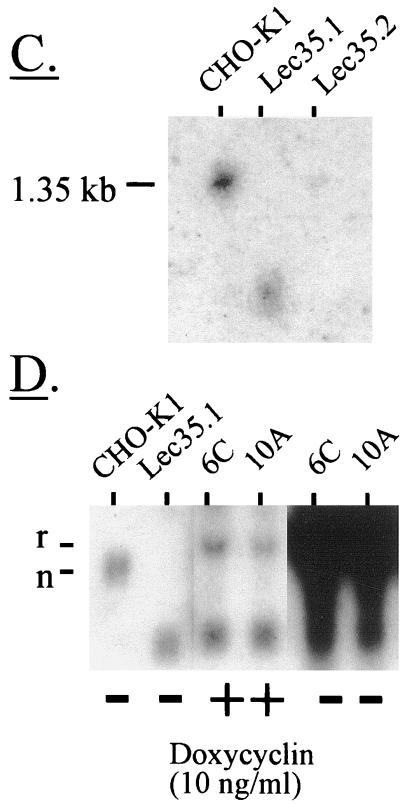
Figure 2.
Analysis of Lec35 nucleic acids. Southern blot analysis of genomic DNA. 32P probes were generated with specific PCR primers and covered either the complete coding sequence of SL15 cDNA (A), or exons 1 and 2, exon 3, exon X, or exons 4–7 (B). Genomic DNA (Easy-DNA; Invitrogen, San Diego, CA) from parental CHO-K1, Lec35.1, or Lec35.2 cells (10 μg for all probes except exon X [50 μg]) was digested with either BamHI or HindIII as indicated and analyzed by Southern blots according to standard methods (Sambrook et al., 1989). The positions of λ phage-HindIII markers are shown. In addition to a strongly hybridizing fragment of ∼6 kbp, a faint BamHI fragment of ∼10 kbp was reproducibly detected with the exon 3 probe in CHO-K1 DNA. Because exon 3 lacks a BamHI site, this is likely to be an irrelevant cross-reacting fragment. Northern blot analysis of RNA. Total RNA was isolated from the CHO-K1, Lec35.1, or Lec35.2 cells lines, as well as the 6C and 10A transfectants of Lec35.1 grown in the absence or presence of 10 ng/ml doxycyclin for 4 d, and 10 μg of total RNA was analyzed by gel electrophoresis and blotting (C and D) as described (Sambrook et al., 1989). A 32P probe prepared from the complete coding sequence of SL15 cDNA was used. Relative intensities of the Lec35 mRNA bands, as described in the text, were measured with a phosphorimager (Fuji, Stamford, CT). The migrations of native (n) and recombinant (r) Lec35 transcripts (D) were reproducible. The position of a 1.35-kb RNA marker (Life Technologies) is shown (C).