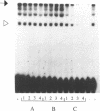
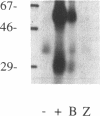
Abstract
An oligodeoxynucleotide that readily flips to the Z-DNA conformation in 10mM MgCl2 was produced by using Klenow enzyme to incorporate 5-bromodeoxycytosine and deoxyguanosine into a (dC-dG)22 template. During synthesis the oligomer can be labeled with 32P to high specific activity. The labeled oligodeoxynucleotide can be used in bandshift experiment to detect proteins that bind Z-DNA. This allows the binding specificity of such proteins to be determined with high reliability using unlabeled linear and supercoiled DNA competitors. In addition, because the radioactive oligodeoxynucleotide contains bromine atoms, DNA-protein complexes can be readily crosslinked using UV light. This allows an estimate to be made of the molecular weight of the proteins that bind to the radioactive probe. Both techniques are demonstrated using a goat polyclonal anti-Z-DNA antiserum.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981 Mar;78(3):1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D. S., Thomas G. H., Elgin S. C. Drosophila nuclear proteins bind to regions of alternating C and T residues in gene promoters. Science. 1989 Sep 29;245(4925):1487–1490. doi: 10.1126/science.2781290. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. The in-vivo occurrence of Z DNA. J Biomol Struct Dyn. 1983 Dec;1(3):593–609. doi: 10.1080/07391102.1983.10507467. [DOI] [PubMed] [Google Scholar]
- Krishna P., Kennedy B. P., Waisman D. M., van de Sande J. H., McGhee J. D. Are many Z-DNA binding proteins actually phospholipid-binding proteins? Proc Natl Acad Sci U S A. 1990 Feb;87(4):1292–1295. doi: 10.1073/pnas.87.4.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafer E. M., Möller A., Nordheim A., Stollar B. D., Rich A. Antibodies specific for left-handed Z-DNA. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3546–3550. doi: 10.1073/pnas.78.6.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meera G., Ramesh N., Brahmachari S. K. Zintrons in rat alpha-lactalbumin gene. FEBS Lett. 1989 Jul 17;251(1-2):245–249. doi: 10.1016/0014-5793(89)81463-2. [DOI] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Sedman S. A., Ebert M. H. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981 Mar 27;211(4489):1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- Möller A., Nordheim A., Kozlowski S. A., Patel D. J., Rich A. Bromination stabilizes poly(dG-dC) in the Z-DNA form under low-salt conditions. Biochemistry. 1984 Jan 3;23(1):54–62. doi: 10.1021/bi00296a009. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Meese K. Topoisomer gel retardation: detection of anti-Z-DNA antibodies bound to Z-DNA within supercoiled DNA minicircles. Nucleic Acids Res. 1988 Jan 11;16(1):21–37. doi: 10.1093/nar/16.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Sheardy R. D. Preliminary spectroscopic characterization of a synthetic DNA oligomer containing a B-Z junction at high salt. Nucleic Acids Res. 1988 Feb 11;16(3):1153–1167. doi: 10.1093/nar/16.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., Sen R., Baltimore D., Sharp P. A. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986 Jan 9;319(6049):154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]