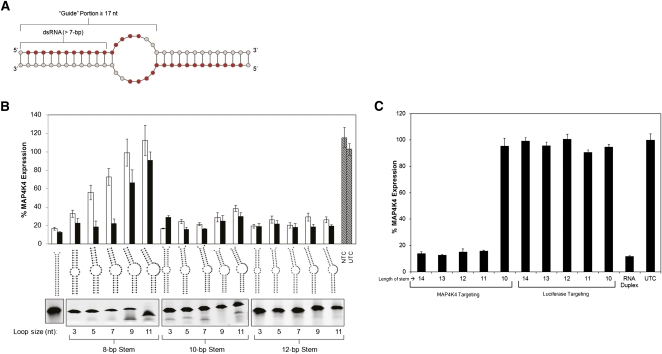
FIGURE 1.
Structural requirements for self-dimerizing single-oligo RISC substrates. (A) 27-nt oligonucleotides were designed to contain a 5′ portion complementary (at least 16 nt, shown in red) to the target mRNA transcript, and a 3′ portion (8–10 nt) enabling oligo self-dimerization into a partially complementary duplex. (B) RNA duplexes composed of two different oligonucleotides (heterodimers that mimic the soloRNA design) were designed and tested in HEK293 cells to investigate the capability of the RNAi machinery to accommodate structurally diverse compounds. The size of the guide strand of the heteroduplex was fixed at 19, 23, and 27 nt for the 8-, 10-, and 12-bp stems, respectively. The loop size was varied through the sequence of the passenger (nontargeting) strand of the duplex. Duplexes were transfected at either 0.5 nM (white bar) or 5 nM (black bar). The panel below the structures depicts a native polyacrylamide gel analyzing the efficiency of the duplex formation. The gel image has been inverted for visual clarity. (C) A panel of soloRNAs (homodimers with stem lengths varying between 10 and 14 bp) targeting MAP4K4 were transfected into HEK293 cells at 1 nM, and gene expression (normalized to PPIB) was evaluated at 48 h. Matching stem length luciferase-targeting soloRNAs were used as negative controls. Untreated cells and a MAP4K4 targeting conventional RNAi duplex (duplex 11546) (Supplemental Table 1) were used as positive controls. Error bars represent the standard deviation of biologic triplicates.