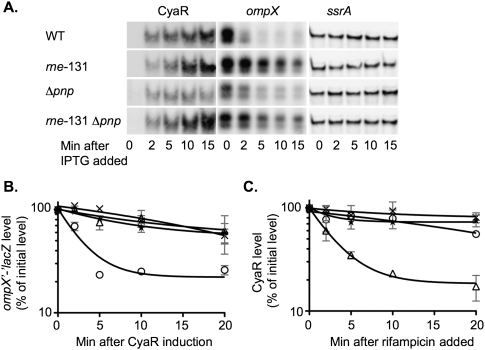
FIGURE 7.
Analysis of the effect of a pnp deletion and the rne-131 allele on CyaR and ompX turnover. (A,B,C) Overnight cultures of the cyaR deletion strain (NRD377; circles or WT), an rne-131 mutant (NRD674; X's or rne-131), a pnp deletion mutant (NRD677; triangles or Δpnp), and an rne-131 Δpnp double mutant (NRD680; stars or Δpnp rne-131) harboring a plasmid expressing CyaR from a lac promoter (pNRD405) were diluted 200-fold in fresh LB containing ampicillin. Cultures were grown at 37°C to an OD600 between 0.3 and 0.4, a sample was removed, and IPTG was then added to each culture. Additional samples were taken 2, 5, 10, and 15 min after IPTG addition. Sixteen minutes after IPTG was added to each culture, rifampicin was added to each culture, and samples were taken 2, 5, 10, and 20 min after rifampicin treatment. The RNA was extracted and processed as described in Figure 4 using the CyaR, ompX, and ssrA probe (A), lacZ probe (B), or CyaR probe (C). The results in A are representative of two experiments. The intensity of the bands in the Northern blots in B and C was quantified using the Multi Gauge software. The band intensity for the 0-min sample was set to 100%, and each additional sample was normalized to the 0-min sample. The results in B and C represent the mean of two experiments, and the standard deviation is indicated by the gray bars.