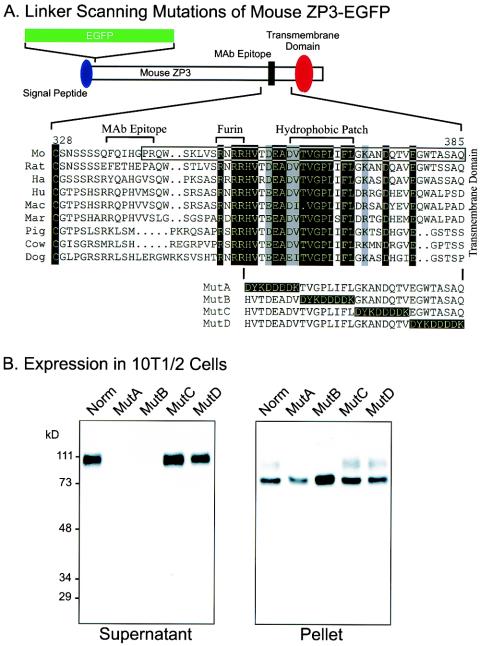
FIG. 2.
Linker-scanning mutations of the carboxyl terminus of ZP3. (A) A second ZP3-EGFP expression vector (top) was used to construct four linker-scanning mutations (MutA, MutB, MutC, and MutD) with a FLAG epitope (DYKDDDDK) to sequentially replace 8-amino-acid domains between the potential proprotein convertase (Furin) cleavage site and the transmembrane domain. Nine amino acid sequences of ZP3 (mouse [Mo], rat, hamster [Ha], human [Hu], macaque [Mac], marmoset [Mar], pig, cow, and dog) were aligned from the terminal, conserved cysteine residue of the zona domain to the predicted transmembrane domain. Residues that are identical in all sequences are indicated as white on a black background; those that were conservative substitutions in all sequences are indicated as black on a gray background. The sequence included in the mouse ZP31-31-EGFP-ZP3342-424aa construct (Fig. 1A) was enclosed in a box. The monoclonal antibody binding site on mouse ZP3 (amino acids 336 to 342), the potential proprotein convertase (furin) recognition site (amino acids 350 to 354), and the hydrophobic patch (amino acids 361 to 369) are indicated above the alignment. (B) Western blot of ZP3-EGFP mutants expressed in heterologous somatic cells. At 3 days after transfection with plasmid vectors encoding either normal or MutA, MutB, MutC, or MutD ZP3-EGFP, 10T[1/2] cell supernatants (left) and pellets (right) were harvested. Proteins in each were separated by SDS-PAGE and transferred to nitrocellulose membranes. Blots were probed with a monoclonal antibody specific to mouse ZP3. Molecular mass markers are indicated on the left in kilodaltons.