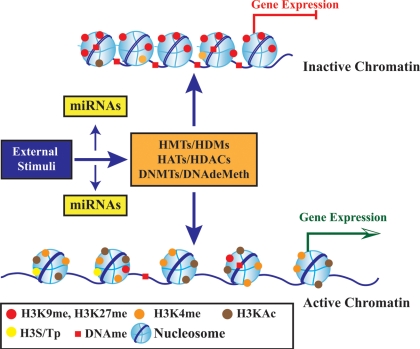
Figure 1.
Schematic diagram showing the role of epigenetic mechanisms in transcription regulation. Chromatin consists of nucleosomes which are made up of DNA wrapped around histone octamers containing dimers of core histone H2A, H2B, H3, and H4. Histone modifications such as H3 lysine-9 methylation (H3K9me) or H3 lysine-27 methylation (H3K27me) are generally repressive marks, whereas H3 lysine-9/14 acetylation (H3KAc) and H3 lysine-4 methylation (H3K4me) are generally activation marks. H3Kme is regulated by histone methyltransferases/demethylases (HMTs/HDMs) and H3KAc is regulated by HATs/HDACs. Histone modifications coupled with DNA methylation at CpG islands by DNMTs/DNA demethylases (DNAdeMeth) determine the active or inactive state of chromatin leading to gene expression or repression, respectively. This dynamic state of the chromatin is subject to alteration by external stimuli via the regulation of epigenetic machinery and miRNAs leading to gene expression and pathophysiological phenotypes.