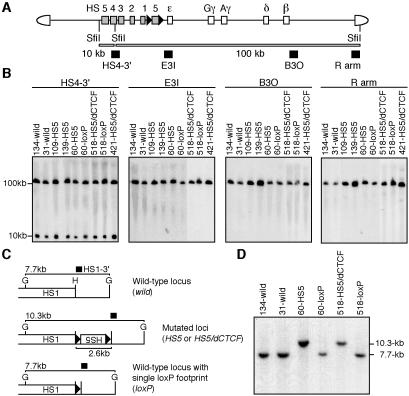
FIG. 2.
Structural analysis of the β-globin YAC in transgenic mice. (A) Schematic representation of the mutant human β-globin YAC. The positions of the β-like globin genes are shown relative to the LCR. SfiI restriction enzyme sites are located 5′ to HS5, between HS4 and HS3, and in the right arm of the YAC. Probes (solid rectangles) used for long-range fragment analysis and expected restriction enzyme fragments with their sizes are shown. (B) Long-range structural analysis of transgenes. The whole β-globin locus is contained within two SfiI fragments (10 and 100 kbp, as in A). DNA from thymus cells was digested with SfiI in agarose plugs, separated by pulsed-field gel electrophoresis, and hybridized separately to probes (shown in A) from the β-globin locus or from the right YAC vector arm. (C) Schematic representation of the transgene locus around HS1. Cre-loxP-mediated HS5 deletion removes the 2.6-kbp insert from the mutated loci (middle), which creates a 7.7-kbp BglII fragment in the locus (bottom). Solid arrowheads, loxP sequences. G, BglII; H, HindIII. (D) Tail DNAs from wild-type (lines 134 and 31), mutant (lines 60-HS5 and 518-HS5/dCTCF), and loxP footprint (lines 60- and 518-loxP) transgenic mouse lines was digested with BglII, separated on an agarose gel, and transferred to a nylon membrane. Hybridization was performed with the HS1-3′ probe (solid rectangle in C). The bands representative of wild-type (7.7 kbp), mutated (10.3 kbp), and footprint (7.7 kbp) loci are indicated.