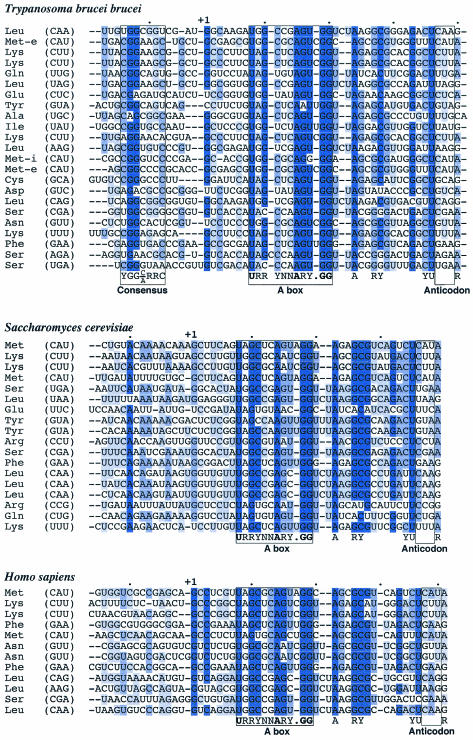
FIG. 1.
Identification of a conserved sequence upstream of T. brucei tRNAs. The coding and upstream sequences of the tRNAs from T. brucei were collected from the website http://zoosun00.unifr.ch/Trypanos/MITBIO.html (30) and from our own work (15). The S. cerevisiae coding and upstream sequences of the tRNAs were collected from the Saccharomyces Genome Database. Human tRNA sequences were extracted from the UCSC Genome Bioinformatics Database using the human BLAST search for sequences with homology to tRNAs of T. brucei. The resulting sequences were analyzed with tRNAScan-SE to predict the presence of tRNA genes (17). The 14-nucleotide upstream sequences plus tRNA coding sequences through position 37 were aligned for T. brucei, Homo sapiens, and S. cerevisiae with Clustal X. The sequences were adjusted manually with MacClade. The conservation at each residue was determined with Jalview. Conserved residues are highlighted as follows: dark blue, >80%; medium blue, >60%; light blue, >40%. The consensus sequence upstream of T. brucei tRNAs, YGG(C/A)RRC, was determined with MacClade. The highly conserved A box for each alignment is boxed, with the consensus sequence below. Invariant residues are shown in bold. Other universally conserved residues are indicated below the alignments. The anticodons for all tRNAs are boxed.