Abstract
Purpose
While lymph node involvement by histopathology informs colorectal cancer prognosis, recurrence in 25% of node-negative patients suggests the presence of occult metastasis. GUCY2C is a marker of colorectal cancer cells that identifies occult nodal metastases associated with recurrence risk. Here, we defined the association of occult tumor burden, quantified by GUCY2C RT-PCR, with outcomes in colorectal cancer.
Experimental Design
Lymph nodes (range: 2–159) from 291 prospectively enrolled node-negative colorectal cancer patients were analyzed by histopathology and GUCY2C quantitative (q)RT-PCR. Participants were followed for a median of 24 months (range: 2–63). Time to recurrence and disease-free survival served as primary and secondary outcomes, respectively. Association of outcomes with prognostic markers, including molecular tumor burden, was estimated by recursive partitioning and Cox models.
Results
In this cohort,176 (60%) patients exhibited low tumor burden (MolLow), and all but 4 remained free of disease (recurrence rate 2.3% [95%CI 0.1–4.5%]). Also, 90 (31%) patients exhibited intermediate tumor burden (MolInt) and 30 (33.3% [23.7%–44.1%]) developed recurrent disease. Further, 25 (9%) patients exhibited high tumor burden (MolHigh), and 17 (68.0% [46.5%–85.1%]) developed recurrent disease (p<0.001). Occult tumor burden was an independent marker of prognosis. MolInt and MolHigh patients exhibited a graded risk of earlier time to recurrence (MolInt, adjusted hazard ratio 25.52 [11.08–143.18]; p<0.001; MolHigh, 65.38 [39.01–676.94]; p<0.001) and reduced disease-free survival (MolInt, 9.77 [6.26–87.26]; p<0.001; MolHigh, 22.97 [21.59–316.16]; p<0.001).
Conclusions
Molecular tumor burden in lymph nodes is independently associated with time to recurrence and disease-free survival in patients with node-negative colorectal cancer.
Keywords: colorectal cancer, molecular staging, occult metastases, tumor burden, prognostic marker, guanylyl cyclase C, quantitative RT-PCR
Regional lymph node metastasis is the single most important prognostic factor in patients with colorectal cancer (1, 2). Although theoretically rendered cancer free by surgery, patients with nodes devoid of histopathologic evidence of cancer (pN0) suffer recurrence rates of approximately 25%, while those rates exceed 50% in patients with ≥4 lymph nodes harboring metastases (pN2) (3, 4). Adjuvant chemotherapy improves disease-free and overall survival in patients with histopathologically evident lymph node metastases, but its role in pN0 patients remains unclear(3, 5–8).
Recurrence in a substantial fraction of node-negative colorectal cancer patients suggests the presence of occult metastases in regional lymph nodes that escape standard detection methods (1, 2). Conversely, patients who are free of lymph node metastases by any detection method may have a better prognosis. Clinically, more accurate assessment of occult metastases would improve risk stratification in a clinically heterogeneous population where up to 25% of patients “cured” by standard care suffer recurrence. In addition, patients with occult metastases at elevated recurrence risk might benefit from the increasingly effective adjuvant chemotherapy available for colorectal cancer.
The intestinal tumor suppressor GUCY2C (guanylyl cyclase C) is the receptor for the paracrine hormones guanylin and uroguanylin, gene products universally lost early in intestinal neoplasia (9, 10). Loss of hormone expression silences GUCY2C signaling which contributes to transformation by promoting proliferation, crypt hypertrophy, metabolic remodeling and genomic instability (10). Highly selective expression by intestinal epithelial cells normally, and universal over-expression by intestinal tumor cells (11–13), suggested that GUCY2C might be a specific molecular marker for metastatic colorectal cancer (14–16). A recent prospective analysis revealed that pN0 colorectal cancer patients whose nodes were GUCY2C positive by molecular analysis suffered recurrence more frequently than those who had GUCY2C-negative nodes (20% v. 6%)(17).
RT-PCR offers a unique opportunity to detect occult tumor cells in lymph nodes(1, 2). In breast and other cancers, the categorical (yes/no) identification of micrometastases is clinically relevant (18). However, due to exquisite sensitivity, RT-PCR can detect cancer cells in lymph nodes below the threshold of prognostic risk (3, 17, 19). Quantitative (q)RT-PCR offers an opportunity to enumerate tumor cells in lymph nodes and by extension, examine the relationship between variable tumor burden and disease risk (19). In addition, qRT-PCR quantifies tumor cells in entire resection specimens (17). Thus, qRT-PCR presents a previously unrecognized method to quantify molecular tumor burden across the regional lymph node network, providing an enhancement over current 2-dimensional histopathology estimates of tumor.
The present analysis defines the association between occult tumor burden in lymph nodes, estimated by GUCY2C qRT-PCR, and time to recurrence and disease-free survival in patients with pN0 colorectal cancer.
Patients and Methods
Study Design
This prospective observational trial at nine centers in the U.S. and Canada explored the prognostic utility of GUCY2C qRT-PCR in lymph nodes of pN0 colorectal cancer patients (17). Investigators and clinical personnel were blinded to results of molecular analyses while laboratory personnel and analysts were blinded to patient and clinical information. To have at least 80% power to detect a hazard ratio of 1.6 (P≤0.05, 2-sided) employing categorical assessment of occult tumor metastases, 225 pN0 patients were required. The study protocol was approved by the Institutional Review Board of each participating hospital. The 291 pN0 patients who met eligibility criteria provided 7,310 lymph nodes (range 2–159, median 21 lymph nodes/patient) for histopathologic examination, of which 2,774 nodes (range 1–87, median 8 lymph nodes/patient) were obtained by fresh dissection and eligible for analysis by qRT-PCR. Disease status, obtained in routine follow-up by treating physicians, was provided for all patients through December 31, 2009.
Patients and Tissues (17)
Between March 2002 and June 2007, we enrolled 299 stage 0–II pN0 colorectal cancer patients who provided informed consent in writing prior to surgery at one of 7 academic medical centers and 2 community hospitals in the U.S. and Canada (Supplementary Fig. 1). Patients were ineligible if they had a previous history of cancer, metachronous extra-intestinal cancer, or perioperative mortality associated with primary resection. For all eligible patients, preoperative and perioperative examinations revealed no evidence of metastatic disease. Lymph nodes, and when available tumor specimens (51%), were dissected from colon and rectum resections and frozen at −80°C within one hour to minimize warm ischemia. Half of each resected lymph node was fixed with formalin and embedded in paraffin for histopathological examination. Lymph node specimens were subjected to molecular analysis if (1) tumor samples, where available, expressed GUCY2C mRNA above background levels in disease-free lymph nodes (>30 copies) and (2) at least one lymph node was provided which yielded RNA of sufficient integrity for analysis (12, 17). Thus, analysis of the 3,093 lymph nodes available from the 299 pN0 patients revealed 236 nodes from 76 patients yielding RNA of insufficient integrity by β-actin qRT-PCR, excluding two patients (Supplementary Fig. 1). Moreover, GUCY2C expression in tumors was below background levels in 6 patients who were excluded from further analysis (12, 17).
RNA Isolation
RNA was extracted from tissues by a modification of the acid guanidinium thiocyanate-phenol-chloroform extraction method (14, 15). Briefly, individual tissues were pulverized in 1.0 mL Tri-Reagent (Molecular Research Center, Cincinnati, OH) with 12–14 sterile 2.5 mm zirconium beads in a bead mill (Biospec, Bartlesville, OK) for 1–2 min. Phase separation was performed with 0.1 mL bichloropropane, and the aqueous phase re-extracted with 0.5 mL chloroform. RNA was precipitated with 50% isopropanol and washed with 70% ethanol. Air-dried RNA was dissolved in water, concentration determined by spectrophotometry, and stored at −80°C.
RT-PCR
GUCY2C mRNA was quantified by RT-PCR employing an established analytically validated assay (12). The EZ RT-PCR kit (Applied Biosystems, Foster City, CA) was employed to amplify GUCY2C mRNA from total RNA in a 50 μL reaction. Optical strip-tubes were used for all reactions, which were conducted in an ABI 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). In addition to the kit components [50 mM Bicine (pH 8.2), 115 mM KOAc, 10 μM EDTA, 60 nM ROX, 8% glycerol, 3 mM Mg(OAc)2, 300 μM each dATP, dCTP, and dGTP, 600 μM dUTP, 0.5 U uracil N-glycosylase, and 5 U rTth DNA polymerase], the reaction master mix contained 900 nM each of forward (ATTCTAGTGGATCTTTTCAATGACCA) and reverse primers (CGTCAGAACAAGGACATTTTTCAT), 200 nM Taqman probe (FAM-TACTTGGAGG-ACAATGTCACAGCCCCTG-TAMRA), and 1 μg RNA template. The housekeeping gene β-actin was amplified employing similar conditions except that forward (CCACACTGTGCCCATCTACG) and reverse (AGGATCTTCATGAGGTAGTCAGTCAG) primers were 300 nM each, while the Taqman probe (FAM-ATGCCC-X(TAMRA)-CCCCCATGCCATCCTGCGTp) was 200 nM. The thermocycler program employed for RT included: 50° × 2 min, 60° × 30 min, 95° × 5 min; and for PCR: 45 cycles of 94° × 20 sec, 62° × 1 min. Reactions were performed at least in duplicate and results averaged.
Statistical Methods
Procedures for reporting statistical methods, including validation procedures for ensuring the accuracy of estimates of hazard ratios and p values, were specifically guided by the REMARK Guidelines (20). Statistical methods for estimating GUCY2C and β-actin mRNA by logistic regression analysis have been described (see Supplementary Information) (17, 21). The primary clinical endpoint was time to recurrence, measured from the date of surgery to the time of the last follow-up, recurrence event or death (17, 22). Disease-free survival, defined as time from surgery to any event regardless of cause, was a secondary outcome (17). Date of recurrence was established by radiographic studies, laboratory studies, physical examination, and/or histopathology. Confidence intervals for raw survival rates were computed by the exact method of Clopper-Pearson (23).
Recursive partitioning, a tree-branching algorithm that identifies homogeneous cohorts in populations, served as the primary analytic approach for survival outcomes, implemented in the R routine RPART (24). This algorithm tests, across all possible variables and levels, for the variable which optimally identifies discrete groups within the study population. The process repeats recursively until a stopping criterion, pre-defined here as the software default of any subgroup with fewer than 20 participants, is achieved (24). Cross-validation (10-fold) during model fitting provided model stability and accuracy and avoided over-fitting. This algorithm was applied using quantitative measures of occult tumor burden as variables for risk stratification. Metrics of occult tumor burden by GUCY2C qRT-PCR included median copy number, maximum copy number, median relative (normalized to β-actin) expression, maximum relative expression, total copy number, and total relative expression across lymph nodes, and the total number of GUCY2C-positive lymph nodes quantified as described (see Supplemental Information) (17, 21). Time to recurrence or disease-free survival served as outcomes in these analyses. Categories of low, medium, and high risk for time to recurrence and disease-free survival were defined by amalgamation (25).
Survival distributions for patients in different risk strata were compared employing the log-rank test. While Kaplan-Meier plots display censored survival at 36 months, analyses incorporated all events up to the date of last follow-up (26). Simultaneous prognostic effects of risk categories and additional covariates were estimated employing Cox regression analysis. Established prognostic variables in the Cox model for recurrence included T stage, grade, lymphovascular invasion, receipt of chemotherapy and/or radiotherapy, anatomical location, number of lymph nodes harvested for histopathology (≥12, <12), and tumor burden risk status defined from recursive partitioning analysis (5). The multivariable model for each outcome included all of the recognized prognostic measures regardless of significance in order to establish the additional independent prognostic effect of occult tumor burden. Since selection of optimal cut-points and subsequent Cox modeling is known to yield inflated alpha level testing (27), 5,000 bootstrap samples were utilized to establish adjusted confidence intervals and empirical p values (28). While comparable as internal validation techniques, bootstrapping is preferred here to cross validation reflecting limitations in populations and events due to cohort segmentation inherent in the latter approach (27). The sensitivity of Cox models employing categorical (yes/no) analysis of occult metastases (17) versus occult tumor burden (how much) were compared using the Akaike Information Criteria (AIC) (29). A global test of proportional hazards for each of the Cox models was completed according to Hosmer and Lemeshow (30). All tests were two-sided, and p<0.05 was considered statistically significant. All analyses were performed with R v 2.9.2, SAS v9.2 and Stata v11.0.
Results
Patient Characteristics
The 291 pN0 patients had a mean age of 68 years (26–90 years) at diagnosis and 55% were male (Table 1). Clinicopathologic features, including depth of tumor penetration (T1/2, T3, T4), and tumor anatomical location (right, left, sigmoid colon) were similar to national experience (3–5). Patients with colon cancer represented 85.9%, while those with rectal tumors comprised 14.1%.
Table 1.
Characteristics of pN0 Patients with Colorectal Cancer
| Variable | Patients | |
|---|---|---|
| N | % | |
| Totals | 291 | 100 |
| Age, years | ||
| <50 | 25 | 8.6 |
| 50–75 | 186 | 63.9 |
| >75 | 80 | 27.5 |
| Sex | ||
| Male | 160 | 55.0 |
| Female | 131 | 45.0 |
| T Stage | ||
| T1/T2 | 120 | 41.2 |
| T3 | 151 | 51.9 |
| T4 | 20 | 6.9 |
| Grade | ||
| Well | 20 | 6.9 |
| Moderate | 226 | 77.7 |
| Poor/unknown | 45 | 15.4 |
| Chemotherapy | ||
| Yes | 65 | 22.3 |
| No | 226 | 77.7 |
| Tumor Site | ||
| Left Colon | 19 | 6.5 |
| Right Colon | 119 | 40.9 |
| Sigmoid Colon | 112 | 38.5 |
| Rectum | 41 | 14.1 |
| Nodes Harvested | ||
| <12 | 49 | 16.5 |
| ≥12 | 242 | 83.5 |
| Lymphovascular Invasion | ||
| No | 233 | 80.1 |
| Yes | 58 | 19.9 |
Occult Tumor Burden and Disease Recurrence
Clinical outcomes in pN0 colorectal cancer patients were analyzed by recursive partitioning using metrics of occult tumor burden estimated by GUCY2C qRT-PCR. The median of relative GUCY2C expression across patient nodes was the dominant quantitative variable stratifying risk. Partitioning algorithms also utilized the maximum relative expression across nodes, the number of positive nodes (17), the median absolute GUCY2C copy number, and the total absolute copy number across nodes to establish risk categories.
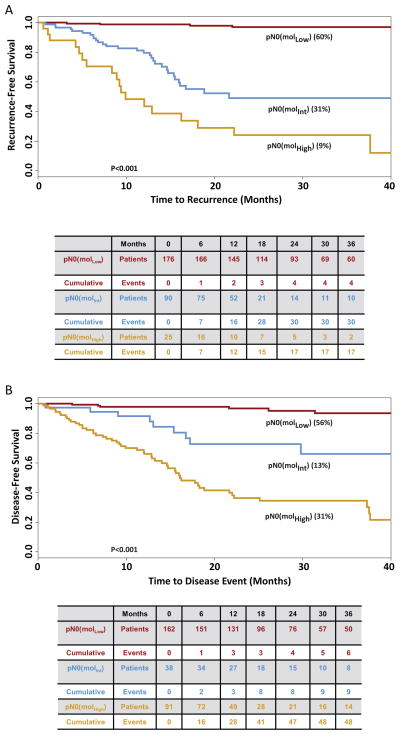
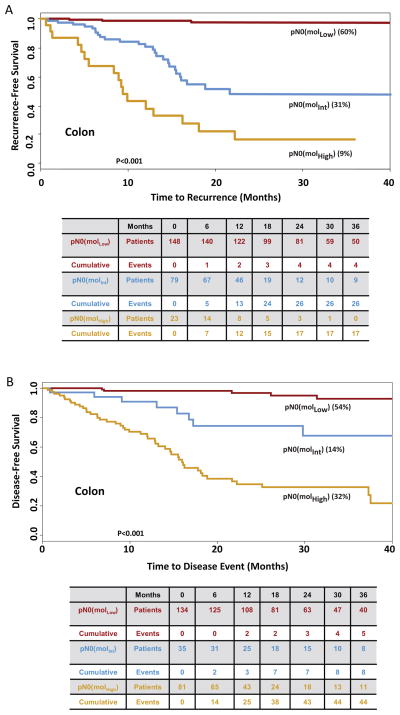
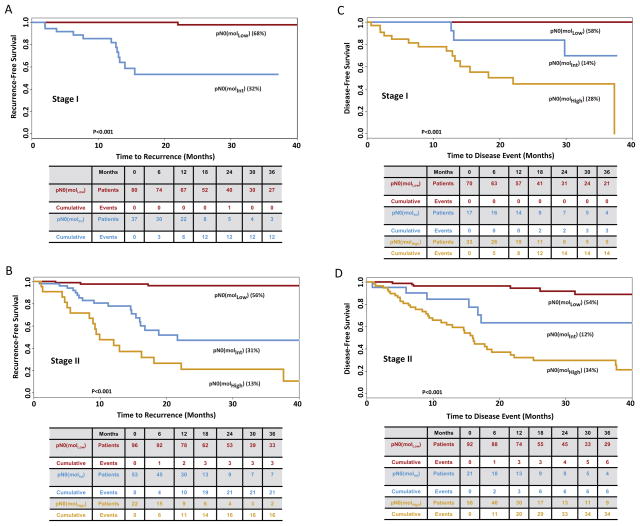
Based on time to recurrence, GUCY2C qRT-PCR stratified pN0 patients into categories in which 176 (60%) patients exhibited low (MolLow), 90 (31%) exhibited intermediate (MolInt), and 25 (9%) exhibited high (MolHigh) (p<0.001) risk of disease recurrence (Fig. 1). Median follow-up was 25 months (range 2–62) for MolLow, 19 months (range 1–61) for MolInt, and 25 months (range 1–63) for MolHigh patients. All but 4 of the MolLow patients remained free of disease during follow-up (recurrence rate 2.3% [95%CI 0.1–4.5%]); 30 (33.0% [23.7%–44.1%]) MolInt patients developed recurrent disease; and 17 (68.0% [46.5%–85.1%]) MolHigh patients developed recurrent disease (p<0.001; Fig. 1). Subgroup analyses revealed that occult tumor burden conferred a substantially worse time to recurrence among patients with colon cancer (Fig. 2), AJCC stage I and II disease (Fig. 3), ≥3 years of follow-up, or optimal collections (≥12) of lymph nodes (Supplemental Fig. 2).
Figure 1. Time to Recurrence (A) and Disease-Free Survival (B) in Patients with pN0 Colorectal Cancer Stratified by Recursive Partitioning.
Tables below Kaplan Meier plots summarize the number of patients at risk as well as cumulative events for each outcome. Censored values in time to recurrence reflect death from another cancer, a non-cancer related death, death due to the cancer treatment, or loss of follow-up of individual patients (22). Censored patients in disease-free survival reflect loss to follow-up (22).
Figure 2. Time to Recurrence (A) and Disease-Free Survival (B) in Patients with pN0 Colon Cancer Stratified by Recursive Partitioning.
Tables below Kaplan Meier plots summarize the number of patients at risk as well as cumulative events for each outcome. Censored values in time to recurrence reflect death from another cancer, a non-cancer related death, death due to the cancer treatment, or loss of follow-up of individual patients (22). Censored patients in disease-free survival reflect loss to follow-up (22).
Figure 3. Time to Recurrence (A, B) and Disease-Free Survival (C, D) in Patients with Stage I (A, C) or II (B, D) Colorectal Cancer Stratified by Recursive Partitioning.
Tables below Kaplan Meier plots summarize the number of patients at risk as well as cumulative events for each outcome. Censored values in time to recurrence reflect death from another cancer, a non-cancer related death, death due to the cancer treatment, or loss of follow-up of individual patients (22). Censored patients in disease-free survival reflect loss to follow-up (22). For the analysis of time to recurrence, there were only 3 Stage I patients stratified as pNo (molHigh). At 6 months, one developed recurrence, one continues to be followed, and one was lost to follow-up.
Similarly, based on disease-free survival, GUCY2C qRT-PCR stratified this population in which 162 (56%) were MolLow, 38 (13%) MolInt, and 91 (31%) MolHigh (<0.001; Fig. 1). For disease-free survival, median follow-up was 24 months (range 2–62) for MolLow, 25 months (range 1–59) for MolInt, and 24 months (range 1 to 63) for MolHigh patients. All but 6 of the MolLow patients remained free of disease during follow-up (3.7% [0.8–6.6%]); 9 (23.7% [6.9–10.2%] MolInt patients developed disease-related events; and 48 (52.8% [42.5–63.0%]) MolHigh patients developed disease-related events (p<0.001; Fig. 1). Like time to recurrence, subgroup analyses suggest that occult tumor burden predicted reduced disease-free survival in patients with colon cancer (Fig. 2), or disease with different stages (Fig. 3), duration or lymph node collections (Supplemental Fig. 2).
Occult Tumor Burden as a Prognostic Variable
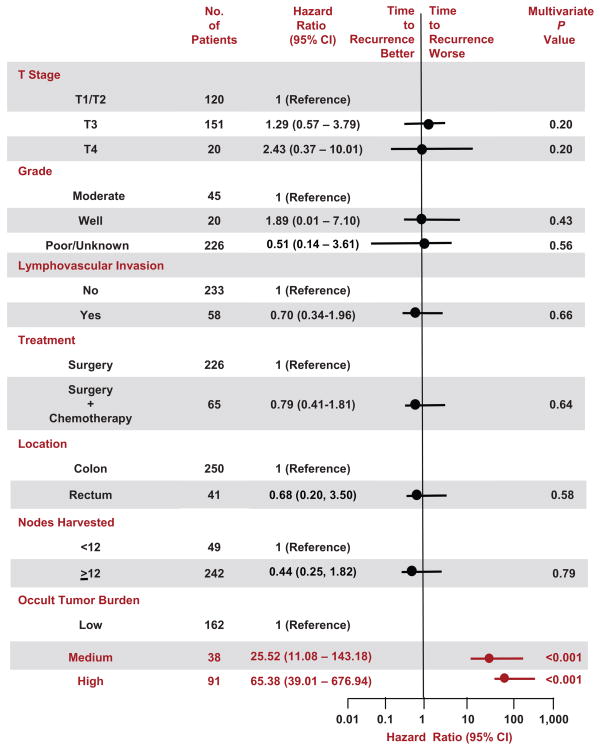
Multivariable analyses employing Cox proportional hazards models (Fig. 4, Supplemental Fig. 3) revealed that canonical prognostic clinicopathologic features contributed little as independent markers of recurrence risk in patients with pN0 colorectal cancer. However, occult tumor burden in lymph nodes provided independent prognostic information. The global test of non-proportional hazards for time to recurrence (chi-square, 6.93; 10 df; p=0.73) and disease-free survival (chi-square, 10.99; 10 df; p=0.36) indicated that there were no significant departures from the proportional hazards assumptions of these models. Patients who were MolInt exhibited time to recurrence (adjusted hazard ratio 25.52 [11.08–143.18]; p=0.001; Fig. 4) and disease-free survival (adjusted hazard ratio 9.77 [6.26–87.26]; p=0.001; Supplemental Fig. 3) comparable to published results for stage III patients (3, 5). Patients who were MolHigh exhibited time to recurrence (adjusted hazard ratio 65.38 [39.01–676.94]; p<0.001; Fig. 4) and disease-free survival (adjusted hazard ratio 22.97 [21.59–316.16]; p<0.001; Supplemental Fig. 3) that approach survival characteristics for patients with stage IV colon cancer (3, 5). Sensitivity analysis revealed that Cox models employing risk categories for time to recurrence established by occult tumor burden were substantially superior (AIC, 470.2) to those employing categorical (yes/no) analysis of occult metastases (AIC, 561.9). Similarly, Cox models employing risk categories for disease-free survival established by occult tumor burden were considerably preferred (AIC, 625.6) over those employing categorical analysis of occult metastases (AIC, 699.7).
Figure 4. Cox Proportional Hazards Analyses of Time to Recurrence in Patients with pN0 Colorectal Cancer Stratified by Recursive Partitioning.
Hazard ratios (circles) with 95% confidence intervals (horizontal lines) and P values for multivariable analyses describe interactions between prognostic characteristics and time to recurrence. Parameters that are significantly prognostic (P<0.05) are highlighted in red.
Discussion
A widely held tenet of cancer staging is the relationship between regional lymph node metastases and prognostic risk (4, 5). In colorectal cancer, lymph node metastasis is the single most important prognostic characteristic, representing pathologic evidence of tumor dissemination beyond its primary location. Clinically, approximately 50% of stage III patients will suffer disease recurrence (1, 2, 4, 5, 8). Because up to 25% of patients with lymph nodes free of tumor involvement also suffer recurrent disease, it is presumed that many such patients harbor occult metastases not identified at the time of primary resection (1, 2).
Under-staging by conventional methods reflects sampling inadequacies inherent in analyzing small volumes of tissue from insufficient lymph node collections (31), and the insensitivity of histopathology, which reliably detects only 1 cancer cell in 200 normal cells (32). Molecular staging can overcome those limitations in the detection of occult lymph node metastases by incorporating all available tissue into analyses, and increasing detection sensitivity through quantifiable, highly sensitive, disease-specific molecular markers(1, 9).
Previously, we demonstrated that prospective, categorical (yes/no) detection of GUCY2C expression in regional lymph nodes was an independent prognostic marker of recurrence risk in pN0 colorectal cancer patients(17). The current results highlight the dramatic enhancement in diagnostic specificity achieved by quantifying molecular tumor burden. When employed as a categorical marker, only 13% of GUCY2C-negative patients were free of occult metastases, but their recurrence risk was low (6%) (17). While recurrence risk was significantly higher (20%) in the 87% of patients who were GUCY2C-positive, most of them did not suffer recurrence (3, 5, 17). It is apparent that nodal metastases, detected by any method, do not assure recurrence; rather they indicate risk. For example, not all stage III patients who by definition have detectable lymph node metastases, ultimately develop recurrent disease (3, 5).
Beyond the categorical presence of metastases, there is an evolving relationship between the quantity of tumor cells in lymph nodes and prognostic risk of recurrence. There is already a well-established correlation between burden of disease, quantified as the number of lymph nodes harboring tumor cells by histopathology, and prognostic risk in colorectal cancer patients. Assuming there are adequate numbers of nodes to review, stage III patients with ≥4 involved lymph nodes exhibit a recurrence rate that is approximately 50–100% greater than those with ≤3 involved nodes(3, 5).
In addition to the number of involved lymph nodes, there is an association between the volume of cancer cells in individual nodes, disease burden, and prognostic risk (3, 31). While metastatic foci ≥0.2 mm are associated with increased disease recurrence, the relationship between individual tumor cells or nests smaller than 0.2 mm and prognostic risk remains undefined (3). The emergence of qRT-PCR provides an unprecedented opportunity for cancer cell enumeration, offering a molecular analogue of the morphological assessment of metastatic volumes by histopathology. Further, quantifying occult metastases in a large volume of tissue (the entire sample), rather than a thin section, and mapping those metastases across the lymph node network enhances 2-dimensional morphology, providing estimates of molecular tumor burden.
Our results suggest that patients with greater occult tumor burden in lymph nodes, estimated by GUCY2C qRT-PCR, have a greater risk of recurrence compared to patients with less tumor burden. In the setting of a common malignancy like colorectal cancer, the quantification of occult tumor burden in lymph nodes to estimate prognostic risk has not been explored previously. Further, the relevant qRT-PCR parameters to estimate tumor burden have not been defined. In the absence of prior experience, recursive partitioning (24) was employed to objectively identify, without bias, parameters that define subgroups of prognostic risk in pN0 patients. Recursive partitioning, applied to all patients using measures of tumor burden established by GUCY2C qRT-PCR stratified pN0 patients into a low risk cohort representing approximately 50–60% of the population, with a very low (<5%) incidence of disease recurrence; an intermediate risk cohort with an incidence of disease recurrence of approximately 33%and a high risk cohort with >60% incidence of recurrence. Multivariable analyses revealed that molecular tumor burden was a powerful independent prognostic marker of time to recurrence and disease-free survival in the context of well-established prognostic clinicopathologic characteristics.
Colon and rectal cancers were considered together in this analysis because GUCY2C is a molecular marker for metastatic tumor cells of intestinal origin and identifies occult tumor burden in patients with either of these diseases (15, 33, 34). While colon cancers were analyzed as a separate cohort, rectal cancer patients were a small minority of the total, providing insufficient numbers for recursive partitioning and risk group analysis. It is noteworthy that the treatment of some rectal cancer patients with neoadjuvant chemoradiotherapy (3, 5, 6) would bias the analysis against the working hypothesis. Indeed, this treatment could produce false negative results, reflecting the absence of adequate lymph node collections for analysis or eradication of occult tumor cells in lymph nodes. However, even in the context of this potential negative bias, the analysis of the full cohort revealed a strong correlation between occult tumor burden and prognostic risk. These results argue for a separate analysis of an adequate population of rectal cancer patients to confirm the utility of occult tumor burden to stratify prognostic risk in those patients.
Tumor burden assessed by GUCY2C qRT-PCR compares favorably with recent gene expression-based efforts to predict colorectal cancer recurrence. Quantification of expression of a 12 gene panel in tumors (Oncotype Dx Colon-Cancer; Genomics Health, Redwood City, CA) stratified 711 stage II (pN0) colon cancer patients into categories in which 40% of patients exhibited a minimum 12% risk, 26% had a maximum 22% risk, while 34% had a risk intermediate between that minimum and maximum, at 36 months (35). Superior specificity, where ~60% of pN0 patients exhibit near-zero risk of recurrence, coupled with a greater demonstrable range of recurrence risk, in the context of a single molecular marker, suggests that quantifying molecular tumor burden by GUCY2C qRT-PCR may offer a diagnostic approach with performance characteristics not previously achieved.
The presence of tumor cells in regional lymph nodes also directs therapy in patients with colon cancer. While adjuvant chemotherapy provides a survival benefit to patients with stage III disease, its utility in patients with pN0 colon cancer remains uncertain, with marginal survival benefits in stage II patients in some, but not all, clinical trials (3, 5–8, 36). This uncertainty of treatment benefit is demonstrated in the evolution of treatment guidelines, in which adjuvant therapy has become discretionary in stage II patients with clinicopathologic features of poor prognostic risk, including T4 stage, intestinal obstruction, and intestinal perforation (8, 36). Heterogeneous responses to therapy in pN0 patients may reflect, in part, the variable presence of occult metastases. Moreover, standard of care includes adjuvant chemotherapy for stage III patients. It is tempting to speculate that MolInt and MolHigh patients, with survival characteristics approximating stage III and IV colon cancer, respectively, might derive benefit from adjuvant therapy. These considerations highlight the importance of advancing beyond the present study to refine the predictive utility of quantifying molecular tumor burden by GUCY2C qRT-PCR. Molecular assessments like GUCY2C analysis could better inform the use of adjuvant chemotherapy in pN0 patients.
In summary, GUCYC2C qRT-PCR analysis of resected lymph nodes in pN0 colorectal cancer patients revealed three discrete strata of recurrence risk ranging from less than 5% to greater than 60%. These results demonstrate, for the first time, the impact of quantitative occult tumor burden estimates on clinical prognosis. They underscore the importance of continuing to validate this novel approach by establishing threshold tumor burden values that can be broadly applied to risk estimation in colorectal cancer patients. Also, they highlight the significance of quantifying the number of lymph nodes required for optimal molecular tumor burden assessment. This molecular approach to occult tumor burden assessment provides a unique opportunity to define the constellation of tumor (microsatellite instability, mutations, methylation, chromosomal instability) and lymph node parameters that optimally estimates prognostic risk of individual patients (37). Moreover, it establishes the importance of defining the contribution of these molecular approaches to therapeutic decision making for node-negative colorectal cancer patients.
Supplementary Material
Acknowledgments
Funding/Support: These studies were supported by grants from NIH (CA75123, CA95026 to SAW and CA112147 to TH) and the Pennsylvania Department of Health and Targeted Diagnostic and Therapeutics Inc. to SAW. SAW is the Samuel M.V. Hamilton Endowed Professor of Thomas Jefferson University.
Footnotes
Disclosure of Potential Conflicts of Interest: SAW is the Chair (uncompensated) of the Scientific Advisory Board of Targeted Diagnostics & Therapeutics, Inc., which provided research funding that, in part, supported this study and which has a license to commercialize inventions related to this work. DSW is a shareholder in Targeted Diagnostics & Therapeutics, Inc.
References
- 1.Iddings D, Ahmad A, Elashoff D, Bilchik A. The prognostic effect of micrometastases in previously staged lymph node negative (N0) colorectal carcinoma: a meta-analysis. Ann Surg Oncol. 2006;13:1386–92. doi: 10.1245/s10434-006-9120-y. [DOI] [PubMed] [Google Scholar]
- 2.Nicastri DG, Doucette JT, Godfrey TE, Hughes SJ. Is occult lymph node disease in colorectal cancer patients clinically significant? A review of the relevant literature. J Mol Diagn. 2007;9:563–71. doi: 10.2353/jmoldx.2007.070032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Compton CC, Greene FL. The staging of colorectal cancer: 2004 and beyond. CA Cancer J Clin. 2004;54:295–308. doi: 10.3322/canjclin.54.6.295. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 5.Greene FL. AJCC Cancer Staging Manual. 6. New York: Springer; 2002. [Google Scholar]
- 6.Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–87. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 7.Quasar Collaborative G, Gray R, Barnwell J, McConkey C, Hills RK, Williams NS, et al. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370:2020–9. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]
- 8.Wolpin BM, Meyerhardt JA, Mamon HJ, Mayer RJ. Adjuvant treatment of colorectal cancer. CA Cancer J Clin. 2007;57:168–85. doi: 10.3322/canjclin.57.3.168. [DOI] [PubMed] [Google Scholar]
- 9.Li P, Schulz S, Bombonati A, Palazzo JP, Hyslop TM, Xu Y, et al. Guanylyl cyclase C suppresses intestinal tumorigenesis by restricting proliferation and maintaining genomic integrity. Gastroenterology. 2007;133:599–607. doi: 10.1053/j.gastro.2007.05.052. [DOI] [PubMed] [Google Scholar]
- 10.Pitari GM, Li P, Lin JE, Zuzga D, Gibbons AV, Snook AE, et al. The paracrine hormone hypothesis of colorectal cancer. Clin Pharmacol Ther. 2007;82:441–7. doi: 10.1038/sj.clpt.6100325. [DOI] [PubMed] [Google Scholar]
- 11.Birbe R, Palazzo JP, Walters R, Weinberg D, Schulz S, Waldman SA. Guanylyl cyclase C is a marker of intestinal metaplasia, dysplasia, and adenocarcinoma of the gastrointestinal tract. Hum Pathol. 2005;36:170–9. doi: 10.1016/j.humpath.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Schulz S, Hyslop T, Haaf J, Bonaccorso C, Nielsen K, Witek ME, et al. A validated quantitative assay to detect occult micrometastases by reverse transcriptase-polymerase chain reaction of guanylyl cyclase C in patients with colorectal cancer. Clin Cancer Res. 2006;12:4545–52. doi: 10.1158/1078-0432.CCR-06-0865. [DOI] [PubMed] [Google Scholar]
- 13.Witek ME, Nielsen K, Walters R, Hyslop T, Palazzo J, Schulz S, et al. The putative tumor suppressor Cdx2 is overexpressed by human colorectal adenocarcinomas. Clin Cancer Res. 2005;11:8549–56. doi: 10.1158/1078-0432.CCR-05-1624. [DOI] [PubMed] [Google Scholar]
- 14.Cagir B, Gelmann A, Park J, Fava T, Tankelevitch A, Bittner EW, et al. Guanylyl cyclase C messenger RNA is a biomarker for recurrent stage II colorectal cancer. Ann Intern Med. 1999;131:805–12. doi: 10.7326/0003-4819-131-11-199912070-00002. [DOI] [PubMed] [Google Scholar]
- 15.Carrithers SL, Barber MT, Biswas S, Parkinson SJ, Park PK, Goldstein SD, et al. Guanylyl cyclase C is a selective marker for metastatic colorectal tumors in human extraintestinal tissues. Proc Natl Acad Sci U S A. 1996;93:14827–32. doi: 10.1073/pnas.93.25.14827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frick GS, Pitari GM, Weinberg DS, Hyslop T, Schulz S, Waldman SA. Guanylyl cyclase C: a molecular marker for staging and postoperative surveillance of patients with colorectal cancer. Expert Rev Mol Diagn. 2005;5:701–13. doi: 10.1586/14737159.5.5.701. [DOI] [PubMed] [Google Scholar]
- 17.Waldman SA, Hyslop T, Schulz S, Barkun A, Nielsen K, Haaf J, et al. Association of GUCY2C expression in lymph nodes with time to recurrence and disease-free survival in pN0 colorectal cancer. JAMA. 2009;301:745–52. doi: 10.1001/jama.2009.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Boer M, van Dijck JAAM, Bult P, Borm GF, Tjan-Heijnen VCG. Breast cancer prognosis and occult lymph node metastases, isolated tumor cells, and micrometastases. J Natl Cancer Inst. 2010;102:410–25. doi: 10.1093/jnci/djq008. [DOI] [PubMed] [Google Scholar]
- 19.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–82. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 20.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–4. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 21.Chervoneva I, Li Y, Iglewicz B, Waldman S, Hyslop T. Relative quantification based on logistic models for individual polymerase chain reactions. Stat Med. 2007;26:5596–611. doi: 10.1002/sim.3127. [DOI] [PubMed] [Google Scholar]
- 22.Punt CJ, Buyse M, Kohne CH, Hohenberger P, Labianca R, Schmoll HJ, et al. Endpoints in adjuvant treatment trials: a systematic review of the literature in colon cancer and proposed definitions for future trials. J Natl Cancer Inst. 2007;99:998–1003. doi: 10.1093/jnci/djm024. [DOI] [PubMed] [Google Scholar]
- 23.Newcombe RG. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998;17:857–72. doi: 10.1002/(sici)1097-0258(19980430)17:8<857::aid-sim777>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 24.Therneau TM, Atkinson EJ. An introduction to recursive partitioning using the rpart routines. Mayo Clinic; 1997. Report No.: Technical report 61. [Google Scholar]
- 25.Sinicrope F, Foster NR, Sargent DJ, Thibodeau SN, Smyrk TC, O’Connell MJ. Model-based prediction of defective DNA mismatch repair using clinicopathological variables in sporadic colon cancer patients. Cancer. 116:1691–8. doi: 10.1002/cncr.24913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pocock SJ, Clayton TC, Altman DG. Survival plots of time-to-event outcomes in clinical trials: good practice and pitfalls. Lancet. 2002;359:1686–9. doi: 10.1016/S0140-6736(02)08594-X. [DOI] [PubMed] [Google Scholar]
- 27.Hollander N, Sauerbrei W, Schumacher M. Confidence intervals for the effect of a prognostic factor after selection of an ‘optimal’ cutpoint. Stat Med. 2004;23:1701–13. doi: 10.1002/sim.1611. [DOI] [PubMed] [Google Scholar]
- 28.Efron B, Tibshirani R, Tibshirani R. An introduction to the bootstrap. Chapman & Hall/CRC; 1993. [Google Scholar]
- 29.Sakamoto Y, Ishiguro M, Kitagawa G. Akaike information criterion statistics. The Netherlands: Dordrecht; 1986. [Google Scholar]
- 30.Hosmer D, Lemeshow S. Applied Survival Analysis: Regression Modeling of Time to Event Data. New York: Wiley; 1999. [Google Scholar]
- 31.Hitchcock CL, Sampsel J, Young DC, Martin EW, Jr, Arnold MW. Limitations with light microscopy in the detection of colorectal cancer cells. Dis Colon Rectum. 1999;42:1046–52. doi: 10.1007/BF02236701. [DOI] [PubMed] [Google Scholar]
- 32.Ratto C, Sofo L, Ippoliti M, Merico M, Bossola M, Vecchio FM, et al. Accurate lymph-node detection in colorectal specimens resected for cancer is of prognostic significance. Dis Colon Rectum. 1999;42:143–54. doi: 10.1007/BF02237119. discussion 54–8. [DOI] [PubMed] [Google Scholar]
- 33.Carrithers SL, Parkinson SJ, Goldstein S, Park P, Robertson DC, Waldman SA. Escherichia coli heat-stable toxin receptors in human colonic tumors. Gastroenterology. 1994;107:1653–61. doi: 10.1016/0016-5085(94)90804-4. [DOI] [PubMed] [Google Scholar]
- 34.Carrithers SL, Parkinson SJ, Goldstein SD, Park PK, Urbanski RW, Waldman SA. Escherichia coli heat-stable enterotoxin receptors. A novel marker for colorectal tumors. Dis Colon Rectum. 1996;39:171–81. doi: 10.1007/BF02068072. [DOI] [PubMed] [Google Scholar]
- 35.Laino C. Gene Test Predicts Return of Colon Cancer. WebMD. 2009 [cited 2009 May 16, 2009]; Available from: http://www.webmd.com/colorectal-cancer/news/20090514/gene-test-predicts-return-of-colon-cancer.
- 36.Benson AB, 3rd, Schrag D, Somerfield MR, Cohen AM, Figueredo AT, Flynn PJ, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22:3408–19. doi: 10.1200/JCO.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 37.Ogino S, Nosho K, Irahara N, Shima K, Baba Y, Kirkner GJ, et al. Negative lymph node count is associated with survival of colorectal cancer patients, independent of tumoral molecular alterations and lymphocytic reaction. Am J Gastroenterol. 2010;105:420–33. doi: 10.1038/ajg.2009.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.