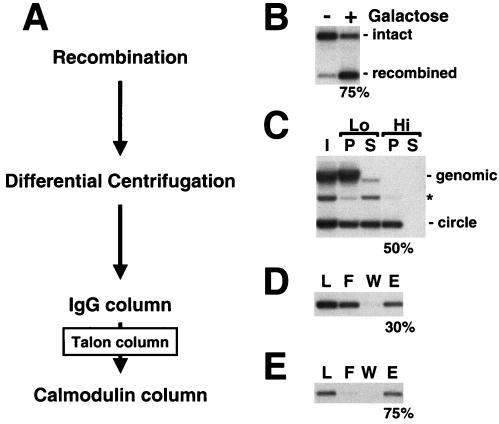
FIG. 2.
(A) Purification scheme for chromatin circles. (B) Site-specific homologous recombination. The PHO5 locus before (−) and after (+) addition of galactose to induce recombinase expression. Genomic DNA was extracted, digested with ApaI and NciI, subjected to 1% agarose gel electrophoresis, and analyzed by blot hybridization with a probe upstream of the PHO5 locus. Positions of the intact and recombined loci are indicated on the right. (C) Differential centrifugation of yeast whole-cell lysates. I, input; P, pellet; S, supernatant; Lo, low-speed spin; Hi, high-speed spin. DNA was extracted, subjected to 1% agarose gel electrophoresis, and analyzed by blot hybridization with a probe specific for the PHO5 locus. Positions of genomic DNA and circle DNA are indicated on the right. The asterisk marks the position of nicked circle DNA that arose as an artifact during DNA preparation. (D and E) IgG and calmodulin affinity chromatography of chromatin circles. L, load; F, flowthrough; W, wash fraction; E, eluate. DNA was analyzed as described for panel C. Numbers on the bottom of each panel give the percentage of recovery of chromatin circles in each purification step, as determined by quantitative blot hybridization.