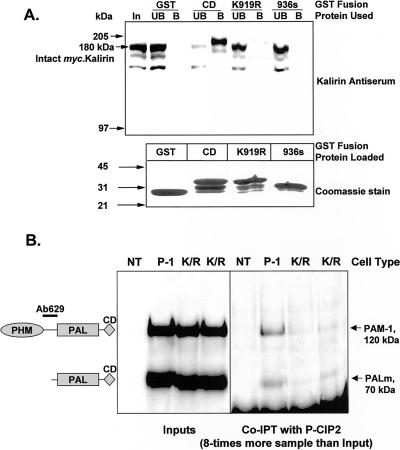
Figure 1.
Verification that mutant PAM-CDs fail to interact with Kalirin or P-CIP2. (A) Kalirin binding to mutant PAM-CD/GST fusion proteins: CHO cells stably expressing myc.Kalirin were extracted and used as the source of Kalirin (Mains et al., 1999). Purified GST, PAM-CD/GST, PAM-CD(K919R)/GST, and PAM-CD936s/GST were each bound to glutathione-Sepharose as described in MATERIALS AND METHODS. The resins were then incubated with CHO cell extract (300 μg of protein) to allow Kalirin binding; unbound proteins were collected by pelleting the resins. The resins were washed, and bound proteins were eluted by boiling in SDS-PAGE sample buffer. Top, comparable aliquots of Kalirin-containing cell extract (In), unbound (UB), and bound (B) fractions were subjected to Western blot analysis using antibody to Kalirin. Bottom, GST-fusion proteins eluted from equal aliquots of the different resins were fractionated by SDS-PAGE and visualized with Coomassie brilliant blue after transfer to a membrane. (B) P-CIP2 binding to mutant PAM-1: nontransfected (NT) AtT-20 cells and AtT-20 cells expressing PAM-1 or PAM-1/K919R (two clones, K/R) were extracted with detergent; Western blot analysis using a PAM antiserum (Ab629) demonstrated equal amounts of PAM protein in the transfected cells (Inputs). Aliquots of each extract were immunoprecipitated with antibody to P-CIP2, fractionated by SDS-PAGE, and visualized with the same PAM antibody (CoIPT with P-CIP2); eightfold more sample was used for coimmunoprecipitation than for analysis of the input. The experiment was repeated three times with the same result; the CoIPT was blocked with excess GST/P-CIP2. The diagram to the left identifies the different PAM proteins and the specificity of Ab629.