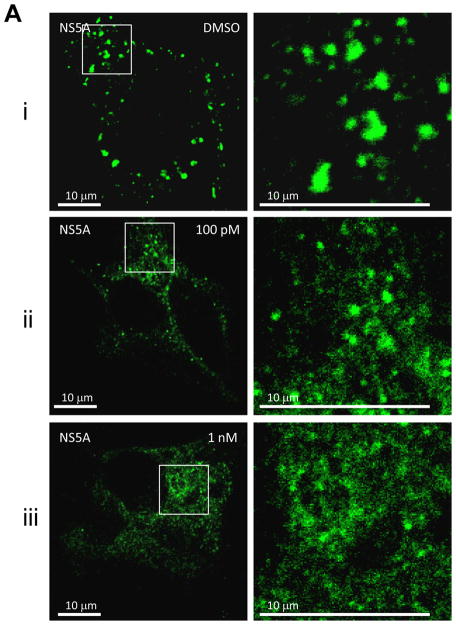
Fig. 2. BMS-790052 alters the subcellular localization of NS viral proteins.
(a) Immunofluorescence analysis of NS5A. Huh7.5 cells were infected with a vaccinia virus expressing a T7 RNA polymerase and then transfected with a plasmid (Bart79I) expressing the HCV NS viral proteins downstream of a T7 promoter. Transfected cells were incubated with 0.1 % of DMSO (i), 100 pM (ii), or 1 nM of BMS-790052 (iii) for 9 hours. Cells were fixed, permeabilized, and stained with a monoclonal anti-NS5A mouse antibody. Anti-mouse Alexa 488-conjugated secondary antibody was used to visualize the NS5A protein in green. White scale bar represents 10 μm. The image on the right panel shows 4-fold enlarged view of the area marked with a white rectangle on the left panel, highlighting the differences in distribution patterns of NS5A.
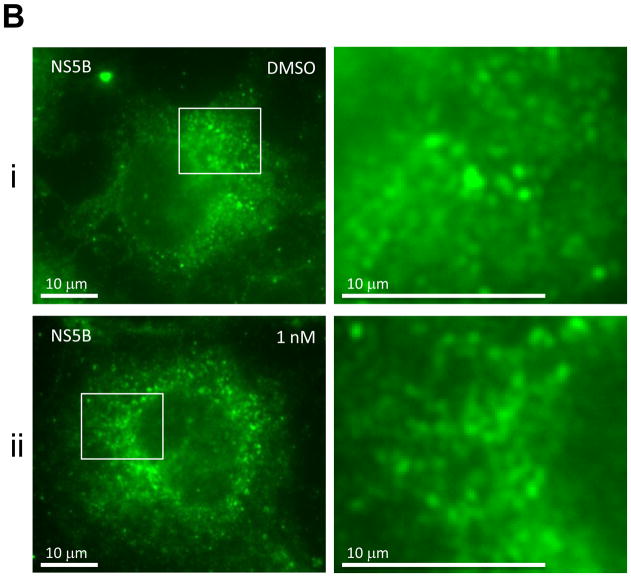
(b) Immunofluorescence analysis of NS5B. Procedure was performed as described in Fig. 2a except that cells were stained with a polycolonal anti-NS5B rabbit antibody. Anti-rabbit Alexa 488-conjugated secondary antibody was used to visualize the NS5B protein in green.
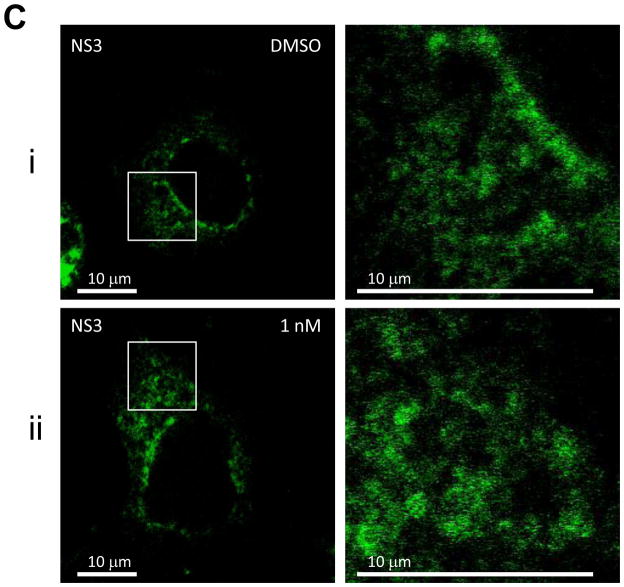
(c) Immunofluorescence analysis of NS3. Procedure was performed as described in Fig. 2a except that cells were stained with a monoclonal anti-NS3 mouse antibody. Anti-mouse Alexa 488-conjugated secondary antibody was used to visualize the NS3 protein in green
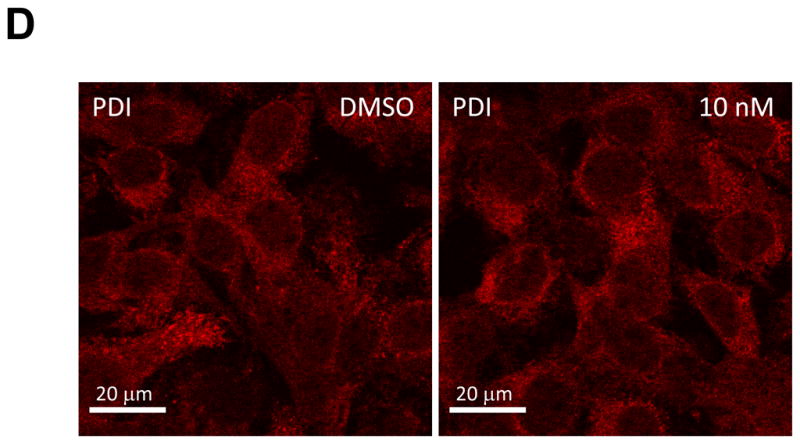
(d) Immunofluorescence analysis of PDI. Procedure was performed as described in Fig. 2a except that cells were stained with a polyclonal anti-PDI rabbit antibody. Anti-rabbit Alexa 594-conjugated secondary antibody was used to visualize the PDI protein in red.