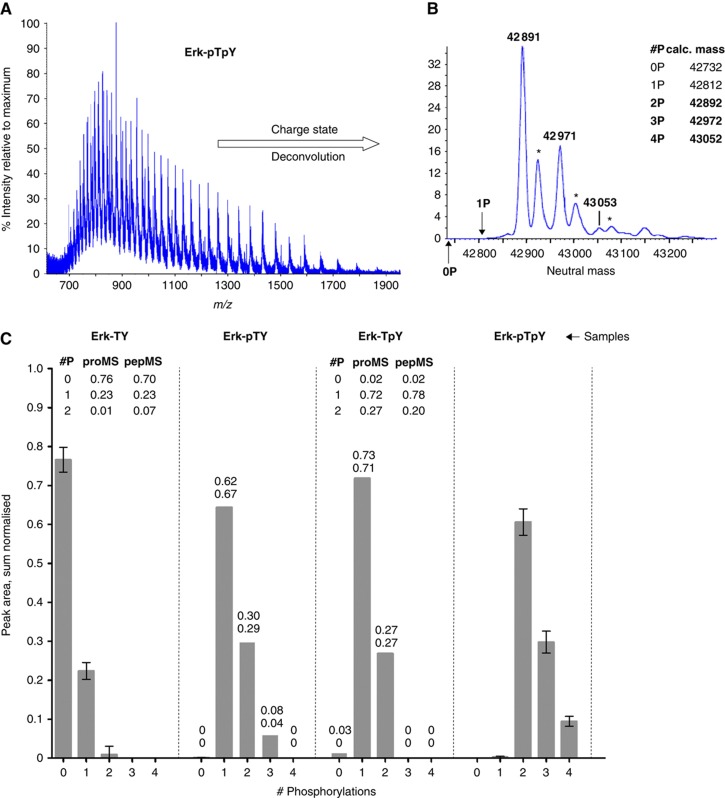
Figure 4.
Protein-based MS analysis of Erk. (A) Mass/charge spectrum of intact Erk-pTpY, showing multiple charge states. (B) Neutral mass spectrum of Erk-pTpY, after charge-state deconvolution. The calculated masses of 15N-labelled His6-tagged Erk2, with 0 to 4 phosphorylations are listed in the inset. Those for 2P, 3P and 4P agree to within 100 p.p.m. with the peaks marked in the spectrum. The positions that would have been occupied by the 0P and 1P states are shown by vertical arrows. Three of the remaining peaks correspond to oxidation products (+32Da, marked by asterisks). (C) Sum-normalised distributions of isobaric groups for each of the four samples, showing 0–4 phosphorylations. Only two phosphorylation sites were detected in the Erk-TY and Erk-TpY samples and the insets compare the proMS values with those calculated from the pepMS data in Figure 2B. Five replicates were used for Erk-TY and Erk-pTpY and the error bars show mean±s.d. Due to insufficient material, only two replicates were possible for Erk-pTY and Erk-TpY. The corresponding columns show the mean, with the two data points listed above to show the variation. Source data is available for this figure at www.nature.com/msb.