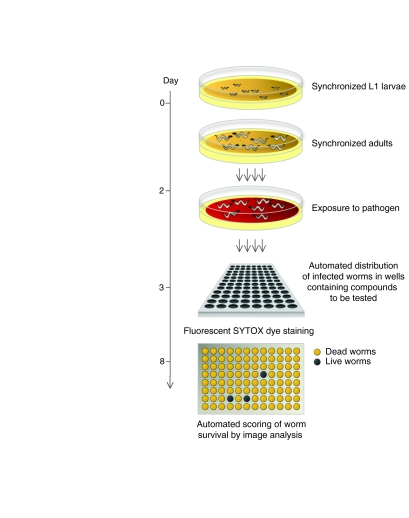
Fig. 2.
A simplified protocol to screen in vivo for new antimicrobial compounds using an established C. elegans infection system and an automated high-throughput assay. After culture and amplification of nematodes on growth plates (seeded with a non-pathogenic E. coli strain), synchronized populations of worms are transferred to infection plates that have been seeded with pathogenic E. faecalis. After half a day, worms are washed off the plates and 15–25 individuals are added to each well of 96-well (shown here) or 384-well microtiter plates that have been pre-loaded with different compounds. After 5 days, the worms in each well are assayed for their survival, using the vital dye SYTOX. Because it is only taken up by dead worms, measuring worm survival can be automated using image analysis software. Wells in which worms live substantially longer contain candidate antibiotics. The figure summarizes the approach taken by Moy et al. (Moy et al., 2009).