Abstract
Unlike the well-characterized nuclear function of the Notch intracellular domain, it has been difficult to identify a nuclear role for the ligands of Notch. Here we provide evidence for the nuclear function of the Notch ligand Delta-like 1 in colon cancer (CC) cells exposed to butyrate. We demonstrate that the intracellular domain of Delta-like 1 (Dll1icd) augments the activity of Wnt signaling-dependent reporters and that of the promoter of the connective tissue growth factor (CTGF) gene. Data suggest that Dll1icd upregulates CTGF promoter activity through both direct and indirect mechanisms. The direct mechanism is supported by co-immunoprecipitation of endogenous Smad2/3 proteins and Dll1, and by chromatin immunoprecipitation analyses that revealed the occupancy of Dll1icd on CTGF promoter sequences containing a Smad binding element. The indirect upregulation of CTGF expression by Dll1 is likely due to the ability of Dll1icd to increase Wnt signaling, a pathway that targets CTGF. CTGF expression is induced in butyrate-treated CC cells, and results from clonal growth assays support a role for CTGF in the cell growth-suppressive role of butyrate. In conclusion, integration of the Notch, Wnt, and TGFbeta/Activin signaling pathways is in part mediated by the interactions of Dll1 with Smad2/3 and Tcf4.
Keywords: colon cancer, Wnt, Notch, Delta like 1, butyrate, Smad, TGFbeta, Activin
Introduction
Canonical Wnt/beta-catenin signaling plays a major role in the development and maintenance of the intestinal architecture and the differentiation of secretory intestinal cells [1]. However, constitutive activation of the pathway, due to mutations in Adenomatous Polyposis Coli (APC), and less frequently, in CTNNB1 and Axin, promotes colorectal carcinogenesis [2–5]. Whereas in most colorectal neoplasms Wnt activity is induced by mutations in its intracellular components, in normal cells Wnt signaling is induced at the plasma membrane level by the binding of Wnt ligands to cell surface receptors. This ligand-receptor binding is transmitted intracellularly, and results in the inhibition of glycogen synthase kinase-3 beta (GSK-3beta). When active, GSK-3beta, in complex with APC and Axin, promotes the phosphorylation and degradation of beta-catenin; however, when GSK-3beta activity is inhibited, dephosphorylated beta-catenin accumulates and interacts with Tcf/Lef DNA-binding proteins. Depending upon the cellular context and the levels of dephosphorylated beta-catenin, the beta-catenin/Tcf transcriptional complexes control the expression of various sets of genes [6,7].
During intestinal development and maintenance, the Wnt pathway interacts with the Notch signaling pathway, which is required for the differentiation of enterocytic intestinal cells [8–9]. The Notch receptor proteins and their ligands (e.g., Delta and Jagged) are cell surface transmembrane molecules. The activation of Notch occurs when its extracellular domain (NECD) interacts with the extracellular domain of a ligand on an adjacent cell; receptor-ligand binding triggers cleavages that release the NECD and the Notch intracellular domain (NICD). The NICD translocates to the nucleus, where it binds to the transcriptional factor CSL, and transactivates promoters with CSL-binding sites (reviewed in [10]).
The Notch and Wnt pathways interact closely in the process of lateral inhibition (11), in which slight variations in the expression levels of Notch and Delta in two adjacent cells are amplified through cell-to-cell interactions. The resulting proteolysis of Notch produces NICD, which suppresses the adoption of a new fate in the cells with higher NICD levels. In the cells with higher Delta levels and lower NICD levels, Wnt signaling promotes the adoption of an alternative cell fate. Thus, goblet cells in the human colon express Delta-like1 (Dll1), and the intracellular domain of this protein may function as a transcriptional regulator promoting the acquisition of a goblet cell phenotype [12]. The process of lateral inhibition, which mediates intestinal cell specification in vivo [9], may also take place to a limited extent in colon cancer (CC) cell populations in vitro. For example, interaction between the Notch and Wnt pathways is likely to maintain the heterogeneity of CC cell populations and colonic neoplasms in terms of Wnt signaling levels. Flow cytometry studies of CC cells expressing the enhanced green fluorescent protein under the control of a Wnt signaling-sensitive promoter have established that the cells respond to butyrate and other histone deacetylase inhibitors (HDACis) as heterogeneous populations, and only a fraction of all cells hyper-activate Wnt signaling [13]. The high or low levels of Wnt activity are not a permanent parameter of specific CC cells within the cell population, since after single cell-sorting each cell regenerates a population with the original heterogeneous character of Wnt activity (unpublished data). Similar heterogeneity of colonic neoplastic cells in terms of Wnt signaling has been demonstrated in vivo [14–17], and this phenomenon represents an obstacle to both cancer prevention and therapy based on compounds that target Wnt signaling. Thus, we have discovered that HDACis hyper-activate Wnt signaling in CC cells, and the fold induction of Wnt transcriptional activity correlates causatively with the apoptotic levels in these cells [13,18–20]. Therefore, in a HDACi-treated cell population, only the cells that undergo high fold changes in Wnt signaling commit to apoptosis; cells that suppress the induction of the pathway are relatively resistant to apoptosis [13,20].
In this report, we demonstrate that exposure of CC cell populations to the HDACi butyrate results in increased levels of phosphorylated Smad2/3 proteins, and that these proteins form complexes with an endogenous Dll1 protein species, and an exogenously expressed Dll1-intracellular domain (Dll1icd). Dll1icd exhibits two activities: (1) it moderately augments Wnt/beta-catenin transcriptional activity, as measured by Wnt signaling reporter systems, and (2) it increases the activity of the endogenous Connective Tissue Growth Factor (CTGF) promoter in butyrate-treated CC cells. The second function of Dll1icd is most likely based upon its ability to associate with the CTGF promoter, as indicated by chromatin immunoprecipitation analyses. A role for CTGF in the high apoptotic response of HCT-116 CC cells to butyrate is supported by results from clonal growth assays. Our findings demonstrate that Dll1icd integrates inputs from the TGFbeta/Activin and Wnt pathways.
Materials and Methods
Cells, plasmids, transfections, luciferase assays, and clonal growth assays
Human CC cell lines were obtained from the American Type Culture Collection (Rockville, MD) and grown in alpha-MEM with 10% fetal bovine serum. The following vectors were provided by various researchers: the pcDNA 3.1-Zeo vector containing the sequence encoding the human Delta-like 1 intracellular fragment, amino acids 569 to 723 (Dr. I. Prudovsky, Maine Medical Center Research Institute, Scarborough, Maine), the human CTGF promoter, −805 to +17 nt (Dr. A. Leask, Schulich School of Medicine and Dentistry, Canada), pTOPFLASH (TOP) and pFOPFLASH (FOP), (Dr. H. Clevers, UMC Utrecht, Utrecht, Netherlands), the mouse Dickkopf1 expression construct (Dkk1) (Dr. D. Wu, Yale University, New Haven, Connecticut), the dominant negative (dn) TCF4 construct (Drs. Bert Vogelstein and Ken Kinzler, Johns Hopkins University, Baltimore, Maryland), and constitutively active Smad2 and Smad3 expression vectors (Dr. D. Danielpour, Case Western Reserve University, Cleveland, OH). The human CTGF promoter was subcloned into the Kpn-XhoI cloning sites of pGL3-Basic luciferase reporter vector (Promega).
Transfections were performed with Lipofectamine 2000 (Life Technologies, Rockville, MD) or via nucleofection with Amaxa (Lonza). We applied the reverse Lipofectamine transfection protocol: complexes between DNA and Lipofectamine are pre-formed in a 96-well plate, and 50,000 cells were added per well. Treatment with sodium butyrate (Sigma, St. Louis, MO) was carried out at 5 mM, and with lithium chloride (Sigma) at 20 mM. Silencing of gene expression was performed with Dll1 ON-TARGETplus SMART pool siRNAS (ThermoFisher), CTGF siRNA (sc-39329, Santa Cruz Biotechnology) and negative control siRNA-A (sc-37007, Santa Cruz Biotechnology). The vector pRSV-TK (Promega Corp., Madison, WI) was used for normalization of transfection efficiency in luciferase reporter assays, which were performed using a Turner Luminometer and a Dual Luciferase kit (Promega, Madison, WI). To evaluate the contribution of TGFbeta/Activin signaling to the effects of butyrate on Wnt, we treated HCT-116 cells transfected with Lef-OT or Lef-OF with an inhibitor of TGFbeta/Activin signaling (SB-505124, Santa Cruz Biotechnology, sc-204341) at 20μM for 24 h.
Clonal growth assays were performed as described previously [13]. For these assays, HCT-116 cells were nucleofected with CTGF siRNAs, and at five hours were mock treated or exposed to 5mM sodium butyrate treatment for 17h. Equal numbers of cells from each treatment were plated in triplicates in 6-well dishes. Ten days later the colonies were stained with crystal violet solution and their numbers determined.
Immunoprecipitations
Nuclear lysates were prepared with the Nuclei EZ kit (N-3408, Sigma). Each immunoprecipitation was carried out with 60–100 μg of nuclear protein and 1–2 μg of normal IgG or Dll1 antibody (sc-8155 or sc-9102, Santa Cruz Biotechnology). After overnight incubation, complexes were precipitated with 20 μl of Protein A/G PLUS-Agarose (sc-2003, Santa Cruz Biotechnology). Four washes were performed with ice-cold phosphate buffered saline; beads were resuspended in 40 μl of Laemmli buffer and protein extracts were analyzed by Western blot analyses.
Chromatin immunoprecipitation (ChIP)
ChIP with antibodies to endogenous Dll1 (sc-8155 and sc-9102, Santa Cruz Biotechnology) were performed with reagents from Santa Cruz Biotechnology and with the EZ-ChIP Chromatin Immunoprecipitation kit (cat. #17-371, Millipore). HCT-116 CC cells were treated for 17h with 5 mM sodium butyrate and cross-linked. Sonicated chromatin containing 60 μg of protein was used for each reaction. The chromatin was pre-cleared for one hour with resin, and then incubated with either 2 μg of normal goat IgG or with 1–2 μg of Delta-like1 antibody (sc-8155, Santa Cruz Biotechnology). Washes and elution of the DNA were performed according to the protocol of the manufacturer. The PCR primers on the CTGF promoter were designed to yield a 143-nucleotide product (5’ primer: 5’-CAATGAGCTGAATGGAGTCCTACA-3’, and 3’ primer: 5’-GTGAGCTGGAGTGTGCCAGC-3’.) PCR conditions were as follows: one cycle at 95°C for 2 min, 50 cycles at 95°C for 1 min, 50°C for 30 seconds, 72°C for 1 min, and extension at 72°C for 5 min. For ChIP with an antibody to the V5 tag of Dll1icd, HCT-116 cells were nucleofected with an empty vector (control) or with a Dll1icd-V5 expression vector. Cells were grown for 48h and treated with 5 mM of sodium butyrate for 17h. The procedure followed the protocol of E. Soler et al. [21].
Western blot analysis
Cytoplasmic-nuclear cell fractionations were performed with the Pierce Nuclear and Cytoplasmic Extraction Reagent Kit (NE-PER). Total cell lysates were obtained as described previously [22]. Equal amounts of protein were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and immunostained with antibodies against Delta-like1 (sc-8155 or sc-9102, Santa Cruz Biotechnology), Activin A (AF338, R&D Systems), anti-V5 (Invitrogen), total Smad2/3 (sc-8332, Santa Cruz Biotechnology) and phospho-Smad2/3 (sc-11769-R, Santa Cruz Biotechnology), CTGF (sc-14939, Santa Cruz Biotechnology), TCF-4 (clone 6H5-3, Millipore, and sc-8632, Santa Cruz Biotechnology), or Actin (A5441, Sigma). Western blots were visualized with a species-specific secondary antibody conjugated to horseradish peroxidase (Sigma) and a chemiluminescence reagent (PerkinElmer Life Sciences, Boston, MA). The specificity of the bands labeled by the goat Dll1 antibody from Santa Cruz Biotechnology (sc-8155) was tested by blocking the antibody with a blocking peptide (sc-8155p) at 1:5 ratio by weight; the Dll1 antibody was used alongside the blocked antibody on duplicate Western blots.
Apoptotic assays
HCT-116 CC cells were plated 24h prior to analyses, and exposed to the agents for 24h. All cells (floating and attached) were harvested and stained for apoptotic and necrotic markers with PE Annexin V Apoptosis Detection Kit I (BD Biosciences, #559763). Flow cytometry analyses were carried out with FACS Aria II and DiVa software. The fold increase in apoptosis was the ratio of percentage of apoptotic cells in butyrate-treated cells to percentage of apoptotic cells in vehicle-treated cells.
Statistics
All P values indicated were calculated using the Student's t-test. Differences were considered significant at P < 0.05.
Results
HCT-116 CC cells exposed to the HDACi butyrate exhibit increased levels of phosphorylated Smad2/3 proteins and nuclear-localized Delta-like 1 (Dll1) protein species
The TGFbeta/Activin pathway, which has been implicated in both the suppression and promotion of carcinogenesis [23], is initiated by the binding of the secreted ligands TGFbeta or Activin A to their cell surface receptors. This binding induces the phosphorylation of Smad2 and Smad3 proteins, which subsequently associate with Smad4, translocate to the nucleus, and activate gene transcription. The TGFbeta/Activin pathway is modified in some, but not all, colon cancers (CCs) via mutation of Smad4, Smad2, TGFbeta receptor (RII), or the Activin A receptor ACVR2 [24–31]. Despite these mutations, there is a significant preservation of TGFbeta/Activin signaling in CC cells, most likely due to the redundancy of TGFbeta/Activin receptors and the existence of Smad4-independent pathways [32–34]. Of particular interest is whether Smad4-independent TGFbeta/Activin signaling exists in CC cells exposed to HDACis (e.g., butyrate), since these agents induce Activin A expression and augment the levels of phosphorylated Smad2 and Smad3 proteins in CC cell lines [35,36]. To evaluate the role of TGFbeta/Activin A signaling in the response of CC cells to butyrate, we have analyzed a pair of CC cell lines: HCT-116 cells, which are characterized by a high level of induction of Wnt activity and apoptosis when exposed to HDACis, and HCT-R cells, which are derived from HCT-116 cells and are resistant to the effects of HDACis [13,18–20]. In agreement with reports on other CC cell lines [35,36], we detected increased levels of phosphorylated Smad2/3, Activin A, and TGFbeta proteins in HCT-116 CC cells exposed to butyrate, but not in HCT-R cells that are resistant to the apoptotic effects of this agent and other HDACis (Fig.1A and B, data not shown). Since exposure to butyrate hyper-induces Wnt signaling and results in apoptosis in CC cells [13,20], we ascertained whether the activation of Smad2/3 proteins contributes to these effects of butyrate by utilizing an inhibitor of Activin receptor-like kinases (ALK) 4, 5, and 7 (SB-505124, Santa Cruz Biotechnology, sc-204341), (Fig.1C). In the absence of butyrate, exposure to the ALK4,5,7-inhibitor did not change significantly Wnt activity: Lef-OT/Lef-OF ratio in vehicle-treated cells was 13.9±3.49, and in inhibitor-treated cells was 15.9±4.56, P>0.05. However, co-treatment of the cells with butyrate and ALK4,5,7-inhibitor decreased the induction of Wnt activity: the Lef-OT/Lef-OF ratio in butyrate-treated cells was 625.0±72.7, and in inhibitor/butyrate-treated cells the ratio was 399.4±79.2, P<0.05. The suppressed induction of Wnt activity in butyrate-treated cells was accompanied by decreased apoptosis, a result that confirms our previous findings for a causal correlation between the fold change of Wnt and the apoptotic levels in butyrate-treated CC cells [13,20]. In the absence of TGFbeta/Activin inhibitor, the fold increase in apoptosis was 4.74±1.02; whereas, in the presence of inhibitor, the fold increase in apoptosis was 2.41±0.62, P<0.05, Fig.1C.
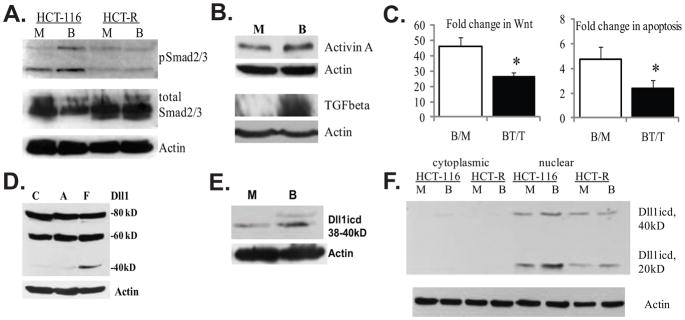
Fig. 1.
Evidence for TGFbeta/Activin signaling and expression of Dll1 species in HCT-116 colon cancer cells. (A). Butyrate induces the phosphorylation of Smad2/3 proteins in butyrate-sensitive HCT-116, but not in butyrate-resistant HCT-R cells. A representative Western blot of total and phosphorylated Smad2/3 in HCT-116 and HCT-R cells exposed to 24h of mock (M) or 5 mM sodium butyrate (B) treatment. The anti-phospho Smad2/3 antibody (sc-11769-R, Santa Cruz Biotechnology) recognizes the phosphorylated forms of both Smad2 and Smad3 (53kD and 60kD). (B). TGFbeta and Activin A expression is augmented in butyrate-treated HCT-116 cells. A representative Western blot analysis of Activin A expression in HCT-116 cells exposed to a 24-h mock (M) or 5 mM of sodium butyrate (B) treatment. Goat anti-Activin A antibody (R&D Systems, #AF338, used at 0.4 μg/ml), recognizes the Activin A dimer (28 kD), consisting of two inhibin beta-chains, as described by others [37]. (C) Suppression of TGFbeta/Activin signaling counteracts the induction of Wnt activity and apoptosis in butyrate-treated HCT-116 cells. Cells were transfected with the Wnt-sensitive luciferase reporters Lef-OT or Lef-OF, and the pRL-null vector, added as a measure for transfection efficiency. Transfected cells were exposed for 24h to mock treatment (M), 5mM butyrate (B), 20μM TGFbeta/Activin inhibitor SB-505124 from Santa Cruz Biotechnology (T), or 20μM inhibitor and 5mM butyrate (BT). Luciferase activity was assayed with dual luciferase kit (Promega), and the ratios of LefOT/LefOF, indicative of Wnt transcriptional activity were calculated. The B/M ratio is the fold increase of Wnt activity in butyrate- vs. mock-treated cells (625.0±72.7/13.9±3.49); whereas the BT/T ratio is the fold increase of Wnt activity in butyrate+inhibitor-treated cells versus inhibitor-treated cells (399.4±79.2/15.9±4.56), P<0.05. The data are the mean of three independent experiments with duplicate samples in each experiment. Apoptosis was measured in cells exposed to the same treatments (see Materials and Methods). The ratio B/M is the ratio of the percentages apoptotic cells in butyrate- vs. mock-treated samples (4.74±1.02); the ratio BT/T is the ratio of the percentages apoptotic cells in butyrate+inhibitor- treated samples vs. inhibitor-treated samples 2.41±0.62, P<0.05. We analyzed three samples per treatment in each of three independent experiments. Bars, SDs. (D). Highest levels of a Dll1icd-containing species are detected in floating fractions of butyrate-treated HCT-116 cells. Exposure of HCT-116 cells to 5 mM sodium butyrate for 48h results in a fraction of floating cells (F) and a fraction of substrate-adherent cells (A). Total protein lysates (150 μg) from these two cell fractions and mock-treated cells (C) were analyzed by Western blotting. The anti-Dll1 antibody detects the full length ligand (80kD), a form of the ligand with partially shed ectodomain (60kD), and a carboxy-terminal form of 38–40kD. (E) Butyrate-treated HCT-116 cells express higher levels of carboxy-terminal 38–40kD Dll1 species compared to mock-treated cells. A representative Western blot analysis of nuclear lysates (80 μg) of HCT-116 cells exposed to mock or 5 mM sodium butyrate treatment for 17h. Detection was achieved with a goat anti-Dll1 antibody (sc-8155, Santa Cruz Biotechnology) and a beta-Actin antibody (Sigma). (F) Highest levels of Dll1icd (20kD) are detected in nuclear fractions of butyrate-treated HCT-116 cells. Cytoplasmic-nuclear fractionation of HCT-116 cells exposed to mock (M) or 5 mM butyrate (B) treatment for 17h was performed with the Pierce Nuclear and Cytoplasmic Extraction Reagent Kit (NE-PER). A total of 100 μg of cytoplasmic or nuclear proteins were analyzed on 12% SDS gels. Western blot detection of Dll1 species was carried out with a rabbit anti-Dll1 antibody (sc-9102, Santa Cruz Biotechnology).
One of the “non-canonical” partners of the Smad2/3 proteins is the intracellular domain of the Notch ligand Delta-like1 (Dll1) [38], and we have observed that HCT-116 CC cells express Dll1 species, including these containing the intracellular domain (Figs.1D–F). In addition to the full-length form of Dll1 (80kD) and a smaller 60kD form, we have observed 38kD and 40kD species that contain the intracellular domain of Dll1 (Dll1icd), Figs. 1D and 1E. These smaller carboxy-terminal forms of Dll1 are better detected in nuclear lysates (Fig.1E), and have been previously reported [39–41]. The 38kD form was enriched in nuclear versus cytoplasmic fractions of mock- and butyrate-treated HCT-116 CC cells (Fig.1F). In the same fractionation experiments, we were able to detect the smallest predicted form of Dll1icd, which is approximately 20kD (Fig.1E). The identity of the protein species recognized by the Dll1 antibody was demonstrated by pre-absorbing the antibody with a blocking peptide (supplementary Fig.1S).
Dll1 associates with Smad2/3 and Tcf4 isoforms in HCT-116 CC cells
Dll1 associates with Smad2/3 in murine neural stem cells [38]. To test whether similar interactions take place in CC cells, we immunoprecipitated the Dll1 protein species from nuclear lysates of mock- and butyrate-treated HCT-116 cells, using an antibody to the carboxy-terminal part of the ligand. These experiments revealed that endogenous Dll1 species co-immunoprecipitate with endogenous Smad2/3 proteins in butyrate-treated cells (Fig.2A). Similar immunoprecipitation experiments also provided evidence for the binding of Dll1 to the transcriptional factor Tcf4, a mediator of Wnt transcriptional activity (Fig.2A). Thus, nuclear lysates of HCT-116 cells immunoprecipitated with Dll1 antibody exhibited augmented presence of low molecular weight forms of Tcf4, and this association was most pronounced in mock-treated cells (Fig.2A). Since Tcf4 has been reported to have multiple alternative splice forms, some of which have a role in the differentiation of gastrointestinal epithelium [42,43], we are currently investigating the precise nature of the Tcf4 isoforms that are immunoprecipitated by the anti-Dll1 antibody. We have consistently detected the low molecular weight forms of Tcf4 in nuclear lysates of HCT-116 cells (Fig. 2B), utilizing two different anti-Tcf4 antibodies (clone 6H5-3, Millipore, and sc-8632, Santa Cruz Biotechnology).
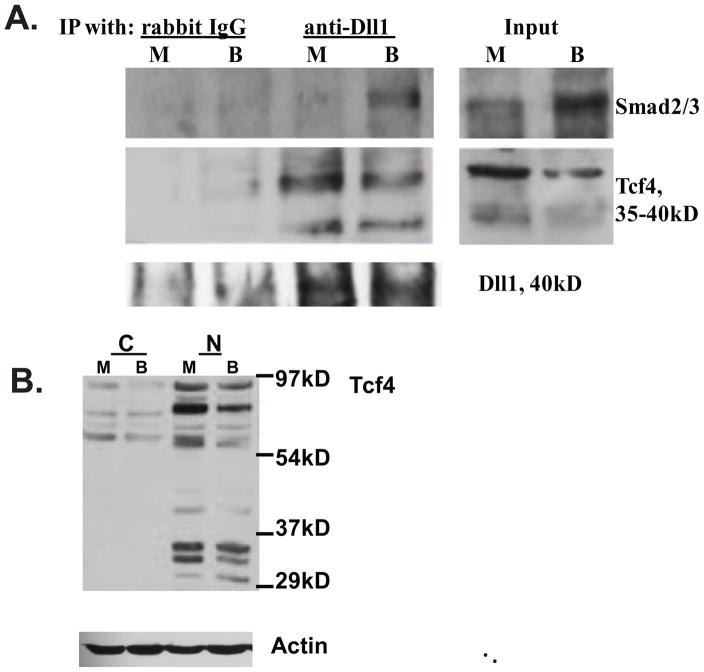
Fig. 2.
Dll1 associates with Smad2/3 and Tcf4 in lysates of HCT-116 CC cells. (A) Immunoprecipitation (IP) analyses with an anti-Dll1 antibody. Nuclear protein lysates (100 μg) from HCT-116 cells, exposed to mock (M) or 5 mM butyrate (B) treatment for 17h, were used for IPs with 2 μg of normal IgG (control) or with 2μg of anti-Dll1 antibody (sc-9102, Santa Cruz Biotechnology). Smad2/3 proteins were detected with a polyclonal antibody that recognizes both proteins (sc-8332, Santa Cruz Biotechnology), Tcf4 was detected with a monoclonal antibody from Millipore (clone 6H5-3). Control experiments confirmed that the anti-Dll1antibody immunoprecipitates the 40kD form of Dll1; we were unable to detect this form in the input, due to its low abundance. (B) Nuclear (N), but not cytoplasmic (C) fractions of HCT-116 cells exhibit the presence of low molecular weight Tcf4 isoforms (30–45 kD). Fractionation of HCT-116 cells exposed to mock (M) or 5 mM butyrate (B) treatment for 24h was performed with the Pierce Nuclear and Cytoplasmic Extraction Reagent Kit (NE-PER). Detection of Tcf4 was carried out as in (A).
Dll1icd enhances Wnt/beta-catenin signaling activity in butyrate-treated HCT-116 cells
We have previously reported the ability of butyrate and other HDACis to upregulate Wnt/beta-catenin transcriptional activity in ten CC cells lines [13,18–20]. Interestingly, Notch ligands are targets of Wnt signaling [44–46]; however, the effects of their intracellular domains on Wnt activity have not been analyzed. Recent data indicate that Dll1icd has transcriptional activity [38,39]; therefore, we tested whether Dll1icd may modulate Wnt signaling levels.
We utilized transient transfection assays to evaluate the effects of exogenously over-expressed Dll1icd (Fig.3A) on the activity of two Wnt signaling reporter systems, TopFlash/FopFlash and Lef-OT/Lef-OF (Figs. 3B and 3C). TOPFlash and Lef-OT contain wild-type Lef/Tcf DNA-binding sites, and thus are Wnt signaling-dependent; FOPFlash and Lef-OF are the corresponding controls, in which the Lef/Tcf DNA-binding sites are modified and not functional. The ratios TopFlash/FopFlash and Lef-OT/Lef-OF are indicative of Wnt-driven transcriptional activity. Wnt activity measured with either of the reporter systems was not changed significantly by exogenous Dll1icd in mock-treated cells: TOP/FOP was 6.6±1.8 and 7.4±1.2 for pcDNA- and Dll1icd-transfected cells, respectively, and Lef-OT/Lef-OF was 6.4±0.2 and 5.4±0.5 for pcDNA- and Dll1icd-transfected cells, respectively. However, in butyrate-treated cells, the exogenous Dll1icd expression increased Wnt levels in a statistically significant manner: TOP/FOP was 35.3±6.6 and 52.4±6.9 (P<0.05) for pcDNA- and Dll1icd-transfected cells, respectively, and Lef-OT/Lef-OF was 16.3±2.7 and 34.8±2.4 (P<0.05) for pcDNA- and Dll1icd-transfected cells, respectively.
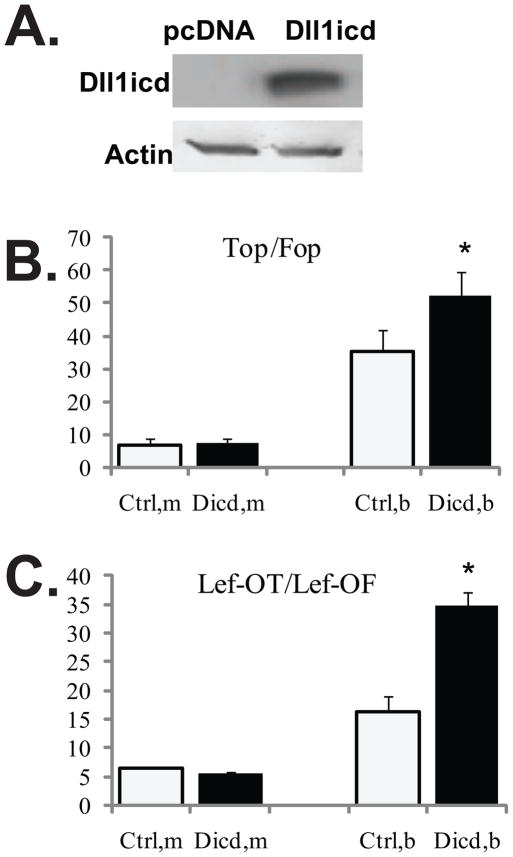
Fig. 3.
The butyrate-induced Wnt activity is augmented by Dll1icd in HCT-116 CC cells. (A) Representative Western blot analyses of HCT-116 CC cells transiently transfected with pcDNA3.1 or a construct expressing Dll1icd. The exogenous protein was detected with a Dll1-specific antibody (sc-8155, Santa Cruz Biotechnology). (B) Dll1icd augments the induction of Wnt signaling by butyrate, as measured with the TopFlash and FopFlash reporter constructs. HCT-116 cells were co-transfected transiently with one of the Wnt signaling reporters (TopFlash or FopFlash), pcDNA3.1 or Dll1icd-expression vector in a 96-well plate. pRL-TK was co-transfected as a measure for transfection efficiency. The ratio between the reporters and the effector vectors was 1:3, the total amount DNA per well was 0.32 μg. Transfections were carried out with Lipofectamine 2000, and treatment with 5 mM butyrate was for 17h. Each transfection was performed in duplicate wells; data represent the mean from results of at least three experiments. Wnt signaling was calculated as the ratio TopFlash/FopFlash (Top/Fop). There was no statistically significant difference between Wnt activity in mock-treated cells transfected with pcDNA3.1 or Dll1icd-expressing construct. For butyrate-treated cells, Top/Fop was 35.3±6.6 and 52.4±6.9 (P<0.05) for pcDNA- and Dll1icd-transfected cells, respectively. (C) Dll1icd augments the induction of Wnt signaling by butyrate, as measured with the Lef-OT and Lef-OF reporter constructs. The transfections were performed as described in 3B. The ratio Lef-OT/Lef-OF is a measure of Wnt activity. There was no significant difference between Wnt levels in mock-treated cells transfected with empty vector or Dll1icd-expressing construct. In butyrate-treated cells, Lef-OT/Lef-OF was 16.3±2.7 and 34.8±2.4 (P<0.05) for pcDNA- and Dll1icd-transfected cells, respectively.
CTGF is an endogenous target of Dll1icd
It has been reported that the intracellular domains of Notch ligands activate gene reporters [38–40,47], and Dll1icd upregulates the endogenous expression of p21 [47]. The transcriptional activity of Dll1icd might be mediated by formation of complexes with the Smad2/3 proteins [38]. During screening for genes that are regulated by butyrate-activated Smad2/3 proteins (Fig.1A), we discovered that butyrate induces the expression of Connective Tissue Growth Factor (CTGF), a secreted matrix-associated protein (Fig.4A).
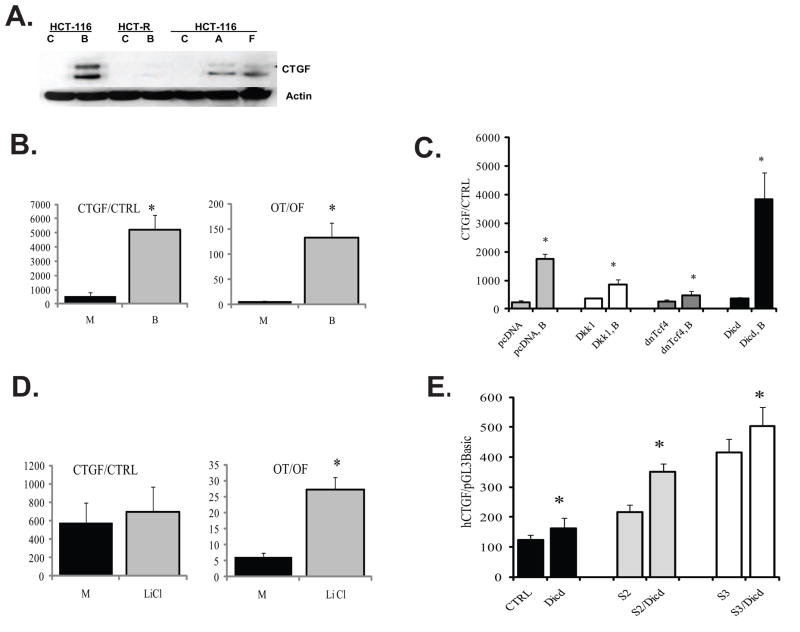
Fig. 4.
Butyrate induces CTGF expression in HCT-116 cells, and this effect is modulated by components of Wnt signaling and Dll1icd. (A) CTGF expression is induced by butyrate in butyrate-sensitive HCT-116 cells, but not in butyrate-resistant HCT-R cells. HCT-116 and HCT-R cells were exposed to mock (C) or 5 mM butyrate (B) treatment for 24h. In addition, HCT-116 cells were treated for 48h with 5 mM butyrate, and lysates from the floating (F) and substrate-adherent (A) cells were analyzed along with lysates of mock-treated (C) cells. Total protein lysates (100μg) were analyzed by Western blot analyses, CTGF and beta-Actin were detected with antibodies from Santa Cruz Biotechnology (sc-14939) and Sigma (A5441), respectively. The production of two CTGF isoforms of 35 and 38 kD, and smaller proteolytically processed forms have been previously reported [48]. (B) Butyrate treatment of HCT-116 cells upregulates the activity of the CTGF promoter. HCT-116 cells were co-transfected with the CTGF reporter and the normalization plasmid pRL-TK, or with the promoterless pGL3Basic and pRL-TK, in a 96-well plate format, following the Lipofectamine 2000 quick transfection protocol (see Materials and Methods). Cells were assayed for luciferase activity after 17-h exposure to mock (M) or 5 mM butyrate (B) treatment. For comparative analyses, the same transfection protocol was applied for Wnt signaling reporters (Lef-OT and Lef-OF). Each transfection was performed in duplicate wells; data represent the mean from results of at least three experiments. (C) Exogenous expression of Dll1icd, dnTcf4, and Dkk1 modulate the induction of CTGF by butyrate. HCT-116 cells were co-transfected with the CTGF promoter reporter (CTGF) or with its promoterless version (CTRL) and one of the following plasmids: pcDNA3Neo (empty vector), Dkk1-expression vector, dnTcf4-expression vector, or Dll1icd (Dicd)-expression vector. All transfections included pRL-TK as a control for transfection efficiency. We applied the quick transfection protocol for Lipofectamine 2000 in 96-well plates. Approximately 60,000 cells were transfected with reporter to effector plasmid ratio at 1:3, to a total of 320ng DNA per well and 0.8 μl of Lipofectamine. After six hours of incubation with the transfection mixture, cells were exposed to mock or 5 mM butyrate treatment for 17h. The ratio of CTGF promoter activity to that of the promoterless reporter was calculated for cells in absence or presence of butyrate. Data represent the mean from results of at least three transfections; each transfection was performed in duplicate wells. There were statistically significant differences between the CTGF promoter activities in butyrate-treated cells expressing Dkk1, dnTcf4, or Dicd and the CTGF promoter activity in butyrate-treated cells transfected with an empty pcDNA vector, (P-values were 0.001, 0, and 0.021, respectively). (D) Increase in active (dephosphorylated) beta-catenin is not sufficient for the induction of CTGF by butyrate. HCT-116 cells were transfected with CTGF or Wnt reporters as described in (B) and exposed to 20mM lithium chloride (LiCl) for 17h. Each transfection was performed in duplicate wells; data represent the mean from results of at least three experiments. There was no statistically significant difference between the activities of the CTGF promoter in mock- and LiCl-treated cells (P>0.05); however, control transfection experiments confirmed the ability of LiCl to induce the Wnt-sensitive promoter Lef-OT (P<0.05). (E) Co-expression of Dll1icd (Dicd) with constitutively active (ca) Smad2 (S2) or constitutively active Smad3 (S3) activates the CTGF promoter in the absence of butyrate. HCT-116 cells were co-transfected with the CTGF promoter reporter (CTGF) or the control pGL3Basic, and one of the following combinations of plasmids: pcDNA3Neo (empty vector), Dll1icd (Dicd)-expression vector, Dll1icd and caSmad2, or Dll1icd and caSmad3. We utilized the reverse Lipofectamine protocol in a 96-well format; the reporters were used at 80 ng per well, the effectors were used at total of 240 ng. pRL-TK was co-transfected at 1:200 ratio for transfection efficiency. Fresh medium was added at five hours post-transfection, and luciferase assays were performed at 24h post-transfection. Data represent the mean from results of at least three transfections; each transfection was performed in duplicate wells. Bars are SDs; statistically significant differences are marked with asterisks.
The increased levels of CTGF protein in butyrate-treated cells is most likely due to the increased gene transcription. Consistent with this, we observed that the exposure of HCT-116 cells to butyrate increases the transcriptional activity of the CTGF promoter (Fig.4B). The increase in CTGF promoter activity is relatively lower than that observed for the Wnt-sensitive reporter Lef-OT, which we have previously shown to be induced by butyrate [13,18–20]. The Lef-OT/Lef-OF ratio was increased by butyrate exposure from 6.6±1.2 to 133.0±29.5 (P<0.05); whereas, the CTGF promoter activity increased from 580±200 to 5,194±1,490 (P<0.05), Fig. 4B.
It has been reported that the CTGF gene is a transcriptional target of TGFbeta/Activin A signaling through Smad- and TGFbeta-response elements in the promoter [49–52]. In agreement with these reports, the induction of CTGF in butyrate-treated HCT-116 cells is concomitant with the phosphorylation of the Smad2/3 proteins by butyrate (Figs.1A and 4A). In addition, butyrate exposure of HCT-R cells, which are resistant to the apoptotic and Wnt activity-inducing effects of butyrate [20] and do not activate the Smad2/3 proteins in the presence of this agent (Fig.1A), does not result in induction of CTGF (Fig.4A). Taking into account the ability of the Smad2/3 proteins to bind Dll1 in HCT-116 cells (Fig.2A), we posited that Dll1 is involved in the transcriptional regulation of CTGF. First, we established that the induction of human CTGF promoter by butyrate in the presence of control plasmid (pcDNA3Neo) was 7-fold: 241±49 in mock-treated cells vs. 1,743±190 in butyrate-treated cells, P<0.05 (Fig.4C). We then determined that the transcriptional induction of the promoter by butyrate was augmented by Dll1icd to 10.3-fold: 369±33 in mock-treated cells versus 3,818±950 in butyrate-treated cells, P<0.05 (Fig.4C). The induction of the CTGF promoter by butyrate was suppressed by dominant negative (dn) Tcf4, a molecule that does not bind to beta-catenin, but binds and blocks Lef/Tcf consensus sites on DNA (Fig.4C). Thus, the induction of CTGF in the presence of dnTcf4 was only 1.8-fold (268±47 in mock-treated cells versus 477±150 in butyrate-treated cells, P<0.05, Fig.4C) compared to the 7-fold induction in the absence of dnTcf4. The validity of these results obtained with a promoterless control vector pGL3Basic (cat. number E1751, Promega) was confirmed in additional experiments that utilized a control luciferase reporter vector with the SV40 promoter (pGL3promoter, cat. number E1761, Promega, supplementary Fig.2S).
Since Tcf4 induces Wnt signaling-dependent promoters in a complex with active beta-catenin, and the activity of the CTGF promoter in some cell types is induced by Wnt ligands [53,54], we analyzed the effects of Dkk1 on CTGF promoter activity. Dkk1 is a secreted protein that blocks the binding of Wnt ligands to their receptors; thus, we have demonstrated that Dkk1 effectively suppresses the induction of Wnt activity in butyrate-treated HCT-116 cells by repressing the activation (dephosphorylation) of beta-catenin [20]. The induction of CTGF promoter activity in the presence of Dkk1 was 2.3-fold (367±24 in mock-treated cells vs. 860±176 in butyrate-treated cells, P<0.005). Therefore, compared to dnTcf4, Dkk1 suppresses the induction of CTGF by butyrate to a lesser extent: in the presence of butyrate, the suppression by dnTcf4 is 3.7±0.8 fold, whereas, that by Dkk1 is 2.3±0.5 fold (Fig.4C).
To further evaluate the potential role of active beta-catenin in the butyrate-induced expression of CTGF, HCT-116 cells were treated with 20 mM lithium chloride (LiCl), a concentration which suppresses glycogen synthase kinase-3beta activity, resulting in increased levels of active (dephosphorylated) beta-catenin [20]. There was no statistically significant difference between the CTGF activity in mock- and LiCl-treated cells (580±200 versus 693±250, P>0.05). In contrast, the same concentration of LiCl induced specifically the expression of the Wnt-sensitive reporter Lef-OT: the Lef-OT/Lef-OF ratio was 6.3±1.3 and 27.1±4.0 in mock- and LiCl-treated cells, respectively (P<0.05). Therefore, whereas LiCl treatment upregulates Wnt/beta-catenin activity more than 4-fold, it does not affect significantly the CTGF promoter activity (Fig. 4D).
Since the inducing effect of Dll1icd on the human CTGF promoter was observed in butyrate- but not in mock-treated HCT-116 cells, we hypothesized that the phosphorylation of Smad2/3 in the presence of butyrate is required for the ability of Dll1icd to induce the promoter. Therefore, we tested whether the CTGF promoter is induced in mock-treated cells by co-expression of Dll1icd with constitutively active (ca) Smad2/3 proteins [55,56]. Co-expression of either ca-Smad2 or ca-Smad3 with Dll1icd resulted in the upregulation of the CTGF promoter activity (Fig.4E). Compared to the transcriptional activity of the CTGF promoter in the presence of a control vector (124±17) or Dll1icd (162±35), the activity of CTGF in the presence of exogenous caSmad2 (216±26), caSmad3 (414±27), or the combinations of caSmad2+Dll1icd (351±47) and caSmad3+Dll1icd (503±64) increased in a statistically significant manner (P<0.05). The increase of CTGF promoter activity in the presence of caSmad3+Dll1icd (503±64) compared to caSmad3 alone (414±27) was not statistically significant; whereas, the increase of CTGF promoter activity in the presence of caSmad2+Dll1icd (351±47) compared to caSmad2 alone (216±26), was statistically significant (P<0.05).
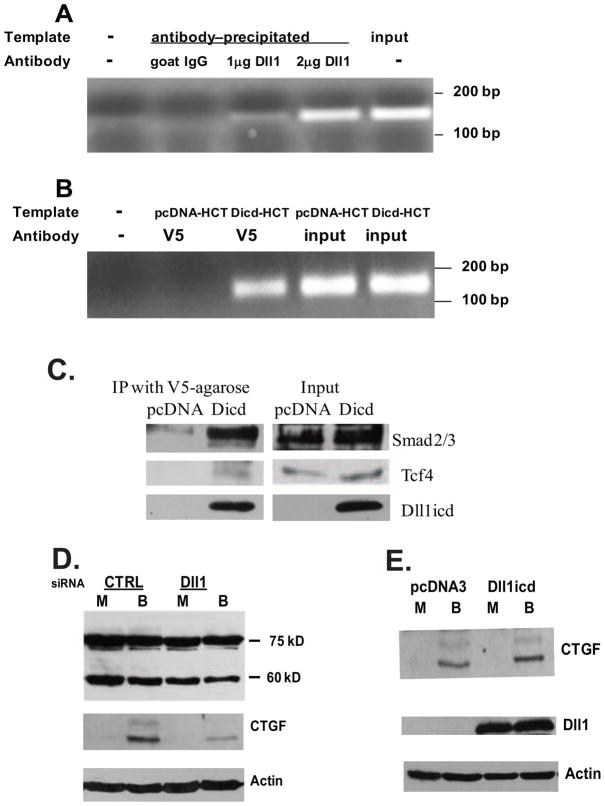
The direct involvement of Dll1icd in the transcriptional control of CTGF was confirmed by (i) chromatin immunoprecipitation (ChIP) analyses of the endogenous CTGF promoter, and (ii) analyses of the CTGF protein levels in cells with modulated Dll1 protein levels. For the ChIP analyses, we first utilized two antibodies to the carboxy-terminal part of Dll1 (sc-8155 and sc-9102, Santa Cruz Biotechnology). Although the results indicated occupancy of a Dll1 protein species on the CTGF promoter (Fig.5A), the performance of these antibodies varied between lots. Since there are no other antibodies to the carboxy-terminal end of Dll1, we undertook an alternative approach by examining the chromatin occupancy of a recombinant Dll1icd tagged with a V5 sequence. In this second series of ChIP experiments, the precipitation step was carried out with anti-V5 beads. Three independent experiments ascertained that exogenous Dll1icd-V5 associates with the CTGF promoter region (Fig.5B). The CTGF promoter region analyzed by these ChIP assays included the Smad-binding and the TGFbeta-response elements, which have been previously reported [49–52]. Western blot analyses of ChIP lysates precipitated with anti-V5 agarose beads revealed the presence of Smad2/3, and Tcf4 (Fig.5C), which is consistent with the immunoprecipitation results obtained with nuclear lysates of untransfected cells (Fig.2A). Finally, moderate downregulation of the endogenous Dll1 expression levels through silencing RNA resulted in a detectable decrease in CTGF protein levels of butyrate-treated HCT-116 cells (Fig.5D); whereas, overexpression of Dll1icd in butyrate-treated HCT-116 cells resulted in a more effective induction of CTGF protein expression (Fig.5E).
Fig. 5.
Dll1icd modulates directly the activity of the CTGF promoter in butyrate-treated HCT-116 cells. (A) Endogenous Dll1 protein species is associated with the CTGF promoter region. Chromatin immunoprecipitation (ChIP) analyses were performed with goat anti-Dll1 or normal goat IgG preparations (Santa Cruz Biotechnology) and lysates of butyrate-treated HCT-116 cells, as described in Materials and Methods. One-fifth of each immunoprecipitation reaction was used as a template for the PCR with CTGF promoter-specific primers. One-half of the PCR products were analyzed on 1.7% agarose gels. On panel A, lane 1 has no DNA template, lane 2 - template precipitated with 2 μg of normal goat IgG, lane 3 - template precipitated with 1 μg of anti-Dll1 antibody, lane 4 - template precipitated with 2 μg of anti-Dll1 antibody, lane 5 – input (0.2% of the input for the ChIP reactions). The PCR product is 143 nucleotides long. (B) Exogenously expressed Dll1icd is associated with the CTGF promoter. ChIP analyses were performed with anti-V5 beads (Sigma) and lysates of butyrate-treated HCT-116 cells nucleofected with an empty vector (pcDNA, control) or with a Dll1icd-expression construct. On panel B, lane 1 has no DNA template, lane 2 - template precipitated from pcDNA-transfected HCT-116 cells, lane 3 - template precipitated from Dicd-transfected HCT-116 cells, lane 4 - template from input, pcDNA-transfected cells, lane 5 - template from input, Dll1icd-transfected cells. (C) Exogenous Dll1icd associates with Smad2/3 and Tcf4 proteins in ChIP lysates of butyrate-treated HCT-116 cells. ChIP lysates from cells transfected with empty vector (pcDNANeo) or Dll1icd-expressing constract were immunoprecipitated with V5-agarose beads (Sigma). The precipitated material was eluted from the beads by adding 2x Laemmli buffer and analyzed via Western blotting. The antibodies for total Smad2/3 and Tcf4 are described in the legend of Fig.2A. (D) Downregulation of endogenous Dll1 levels results in a suppressed induction of CTGF. Nucleofection of HCT-116 cells was carried out as described in Materials and Methods. Cells were exposed to mock treatment or 5 mM butyrate for 17h and total cell lysates were analyzed by Western blotting with a CTGF antibody. (E). Overexpression of Dll1icd in HCT-116 cells results in enhanced induction of CTGF. Dll1icd was overexpressed in HCT-116 cells were nucleofected with pcDNA or Dll1icd expression vector, exposed to mock or 5 mM butyrate for 17h, and total cell lysates were analyzed via Western blotting for CTGF protein expression. The signal for beta-Actin was used as a loading control.
Suppression of CTGF expression promotes the survival of HCT-116 cells exposed to butyrate
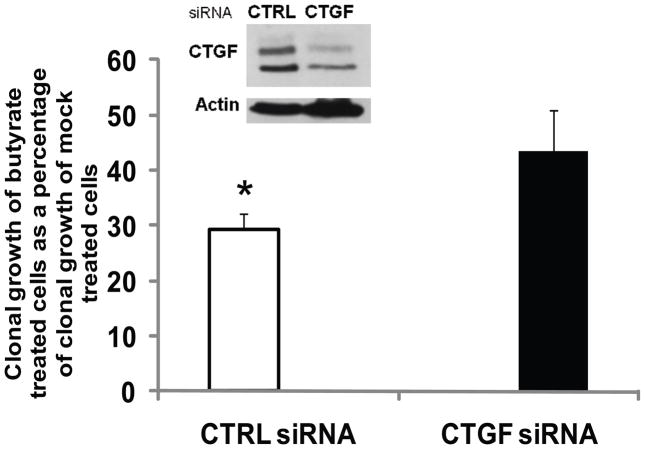
To evaluate the physiological significance of the butyrate-induced CTGF in CC cells, we suppressed CTGF expression with specific siRNA introduced via nucleofection. The nucleofection typically results in more than 80% transfection efficiency in HCT-116 cells, as established by transfections with a GFP-expressing reporter vector (data not shown); however, the maximal downregulation of CTGF expression was ~50% as estimated with Western blot analyses (Fig.6, inset). Clonal growth assays with cells nucleofected with control or CTGF siRNA established that the butyrate-induced CTGF expression contributes to the sensitivity of HCT-116 CC cells to the growth suppressive effects of butyrate (Fig.6). Thus, cells transfected with control siRNA and exposed to butyrate (5 mM, for 17 hours) exhibited a level of clonal growth 29.2% of that observed in control siRNA-transfected cells exposed to mock treatment. However, cells transfected with CTGF siRNA and exposed to butyrate (5 mM, 17 hours) exhibited a level of clonal growth 43.4% of that observed for CTGF siRNA-transfected cells that were mock treated (Fig.6). Therefore, the suppressed CTGF protein levels augmented the resistance of HCT-116 cells to the growth-inhibitory effects of butyrate.
Fig. 6.
Suppressed CTGF expression contributes to the resistance of HCT-116 cells to the growth inhibitory effects of butyrate. HCT-116 cells (106) were nucleofected with 175 pmol of control (non-specific) or human CTGF siRNA (Santa Cruz Biotechnology) and incubated in two wells of a 6-well plate. At six hours post-transfection the cells were mock-treated or exposed to 5 mM sodium butyrate for 17 hours. The inset is a representative Western blot analysis of the CTGF protein levels in control and CTGF siRNA-nucleofected cells exposed to 5 mM butyrate for 17h. Clonal growth assays were performed as described in Materials and Methods. The clonal growth of butyrate-treated cells is expressed as a percentage of the clonal growth of the mock-treated cells. The clonal growth assays were repeated at least three to six times. Statistically significant differences are marked with asterisks. Bars, SDs.
Discussion
A nuclear role for the intracellular domains of Notch ligands, including Dll1, has been reported by several research groups [38–41,47,57] and reviewed by D’Souza et al. [58]. The nuclear localization of Dll1icd is likely due to two motifs of positively charged amino acids, since mutations in these amino acids impair the nuclear translocation of the protein [47]. We have observed that butyrate treatment increases the steady-state levels of Dll1icd in nuclear fractions of HCT-116 colon cancer cells (Fig.1F).
The Notch ligands Dll1 and Jagged1 are induced by Wnt/beta-catenin signaling [44–46]. However, our data suggest a positive feedback in butyrate-treated HCT-116 CC cells, since the intracellular domain of Dll1 augments the induction of Wnt transcriptional activity in these cells (Fig.3). The fold induction of Wnt activity by butyrate was increased moderately in the presence of Dll1icd: 1.5-fold, when measured with the TopFlash and FopFlash reporters, and 2.2-fold, when measured with the Lef-OT and Lef-OF reporters. The increase was due to the augmented Wnt transcriptional activity in butyrate-treated cells. These findings, combined with the observation that Dll1 species co-immunoprecipitate with Smad2/3 and Tcf4 proteins (Fig. 2), suggest that Dll1icd may affect Wnt activity through various mechanisms. Since the binding between Tcf4 and Dll1 protein species is strongest in mock-treated HCT-116 cells, it is possible that in these cells, Dll1 participates in repressive transcriptional complexes with Tcf4. However, in butyrate-treated HCT-116 cells, Dll1 species may bind phosphorylated Smad2/3 proteins to form activating transcriptional complexes. A direct induction of Wnt transcriptional reporters by Smad2/3 and a Lef/Tcf protein has been reported [56,57]; therefore, it is possible that complexes between Dllicd1, Smad2/3, and Tcf4 directly modulate some Wnt-targeted promoters through their Lef/Tcf DNA-binding sites. Alternatively, the effect of Dll1icd on Wnt activity might be indirect; complexes between Dll1icd and phosphorylated Smad2/3 proteins may target only genes with Smad-binding elements [38]; subsequently, the TGFbeta/Activin transcriptional activity can alter the expression of modulators of Wnt activity. Thus, microarray analyses have revealed that TGFbeta/Activin signaling augments the expression of Wnt receptors, and suppresses the expression of Wnt signaling inhibitors [59]. Regardless of the mechanism, we have shown that the pharmacological inhibition of TGFbeta/Activin signaling decreases Wnt activity and Wnt-dependent apoptosis in butyrate-treated HCT-116 CC cells (Fig.1C), demonstrating a cooperation between TGFbeta/Activin and Wnt signaling in the effects of a histone deacetylase inhibitor such as butyrate.
The induction of the CTGF promoter by Dll1icd (Fig.4) may also be accomplished by direct and indirect mechanisms, which are not mutually exclusive. The direct mechanism is supported by our finding of the chromatin occupancy of the CTGF promoter by Dll1icd (Figs. 5A and 5B). The association of Dll1icd with the CTGF promoter sequence is likely mediated by Smad2/3 proteins, which target the Smad-binding element (SBE) on the CTGF promoter [49–52]. Corroborating this hypothesis is our finding that both endogenous Dll1 species and recombinant Dll1icd co-immunoprecipitate with endogenous Smad2/3 proteins from nuclear lysates of butyrate-treated HCT-116 cells (Fig.2A and 5C). These findings are in agreement with the report of Hiratochi et al. that SBEs are a prime target of Dll1icd [38]. The transcriptional function of Dll1icd through SBEs may not be unique to the CTGF promoter; thus, p21, the first reported endogenous target of Dll1icd [47], is also a TGFbeta-regulated gene with imperfect matches of the SBE and TGFbeta-response element within its regulatory regions [60,61]. Therefore, it will be of interest to determine whether Dll1icd and Smad2/3 cooperatively induce the p21 promoter.
We have demonstrated that Dll1 protein species co-immunoprecipitate with short Tcf4 isoforms (Figs.2A and 5C). The levels of Tcf4 immunoprecipitated by anti-Dll1 antibody decrease in butyrate- versus mock-treated HCT-116 cells (Fig.2A); however, the association between Dll1icd and Tcf4 species in nuclear lysates from butyrate-treated cells was confirmed by immunoprecipitations with ChIP lysates from Dll1icd-transfected cells (Fig.5C). The decreased binding of endogenous Dll1 species to Tcf4 in butyrate-treated cells might be due to the decreased steady-state levels of Tcf4 [Fig.2B and ref.20]. Since dnTcf4 (Fig.4C) and full length Tcf4 (data not shown) decrease the induction of the CTGF promoter activity by butyrate, it is possible that Dll1icd-Tcf4 complexes act as transcriptional repressors of CTGF and other genes in the absence of phosphorylated Smad2/3 proteins (as in mock-treated HCT-116 cells). Such transcriptional repression may occur in differentiated intestinal cells; thus, Tcf4 is expressed at the highest levels in non-cycling differentiated cells of the colonic epithelium [62,63]. Interestingly, Nawshad et al. have reported that Lef1, another Lef/Tcf factor, forms transcriptionally repressive complexes with Smad2 and Smad4 [64]; therefore, the repressive complexes of Dll1icd and Tcf4 may include non-phosphorylated Smad2/3 proteins. Butyrate treatment may promote the transition from repressive to activating Dll1icd-complexes by (i) decreasing the steady-state levels of Tcf4 protein (see Fig.2B and ref.20), (ii) titrating Tcf4 away from Dll1icd due to increased binding of Tcf4 to beta-catenin [20], and (iii) increasing the levels of phopshorylated Smad2/3 proteins.
The indirect mechanism, by which Dll1icd induces the CTGF promoter activity, is likely based upon the ability of Dll1icd to moderately enhance Wnt signaling (Fig.3). Thus, the ability of Wnt signaling to maintain and/or upregulate the expression of CTGF has been observed in different experimental models [65,66], and Wnt10b and Wnt3 ligands upregulate the CTGF promoter activity in NIH 3T3 fibroblasts via the SBE [53,54]. We demonstrated that Dkk1, a molecule that inhibits the activation (dephosphorylation) of beta-catenin in butyrate-treated CC cells [20], moderately suppresses the induction of CTGF promoter by butyrate. However, lithium chloride, which activates beta-catenin, is unable to induce the CTGF promoter (Fig.4C and D); therefore, Wnt transcriptional complexes between active beta-catenin and Tcf4 are insufficient for the direct induction of CTGF. Compared to Dkk1, dnTcf4, another inhibitor of Wnt signaling, more effectively suppressed the induction of CTGF by butyrate (Fig. 4C); however, as discussed above, exogenous full length Tcf4 mimics the effect of dnTcf4 on the CTGF promoter (data not shown). Therefore, dnTcf4 most likely suppresses the CTGF promoter activity not by downregulating Wnt signaling, but by binding Dll1icd, and/or non-phosphorylated Smad2/3. This possibility is in agreement with the report of Labbe et al. that the activity of Smad3-Lef1 complexes requires Smad- and Lef/Tcf DNA-binding elements, but is independent of beta-catenin [67]. Although we were unable to detect immunoprecipitation of beta-catenin by anti-Dll1 antibody (data not shown), it is possible that under different conditions beta-catenin participates in a complex with Smad2/3 proteins and Tcf4 [68,69].
Consistent with a role of Dll1icd in the induction of CTGF expression by butyrate, moderate modulation of Dll1 expression resulted in corresponding changes in CTGF protein levels. Thus, a moderate suppression of endogenous Dll1 expression resulted in a decreased induction of CTGF by butyrate (Fig.5D); whereas, overexpression of recombinant Dll1icd resulted in detectable upregulation of the CTGF induction by butyrate (Fig.5E). The modest effects of Dll1 modulation on CTGF protein levels may be due to the fact that the cells already express sufficient amounts of endogenous Dll1icd (Fig.1E, nuclear fraction of butyrate-treated HCT-116), and the levels of phosphorylated Smad2/3 proteins is a limiting factor.
Evidence supporting transcriptional cooperation between the TGFbeta/Activin and Wnt signaling pathways was first provided by an oligonucleotide microarray assay that identified common Wnt and TGFbeta target genes in normal murine mammary gland epithelial cells [70]. The combined treatment of these cells with TGFbeta and Wnt revealed a novel transcriptional program that differed from that of single ligand treatments; interestingly, among the cohort of genes that were cooperatively induced by both pathways was the murine Ctgf gene. Based upon our findings, we propose that cooperativity between TGFbeta/Activin and Wnt signaling pathways is in part mediated by the intracellular domain of Delta-like 1, and its association with Tcf4 and Smad2/3.
CTGF has been reported to increase the migration and invasion of some types of cancer cells [71–74]; however, there are also reports that CTGF inhibits cancer metastasis and invasion [75], acts as a tumor suppressor [76], and contributes to apoptosis of various cell types [77–79]. The dual function of CTGF in proliferation and apoptosis of cancer cells [52] prompted us to determine the role of CTGF induction in butyrate-treated HCT-116 cells. We have previously reported that among ten CC cell lines, HCT-116 cells exhibit one of the highest sensitivities to the growth inhibitory and apoptotic effects of butyrate [13]. The role of CTGF in the response of HCT-116 CC cells to butyrate was evaluated by clonal growth assays of cells with suppressed CTGF expression. Moderate silencing of CTGF expression in HCT-116 cells was accompanied by decreased sensitivity of the cells to the growth inhibitory effect of butyrate (Fig.6). In agreement with the role of CTGF in mediating butyrate-induced cell growth suppression, HCT-R cells, which are resistant to the apoptotic and growth-suppressive effects of butyrate [20], exhibit suppressed induction of CTGF in the presence of butyrate (Fig.4A). Although our findings suggest that CTGF contributes to the growth suppressive effect of butyrate in HCT-116 CC cells, we have not observed induction of CTGF by butyrate in other butyrate-sensitive CC cell lines (data not shown); therefore, the cell growth-suppressive effects of butyrate take place even in the absence of CTGF expression.
Conclusions
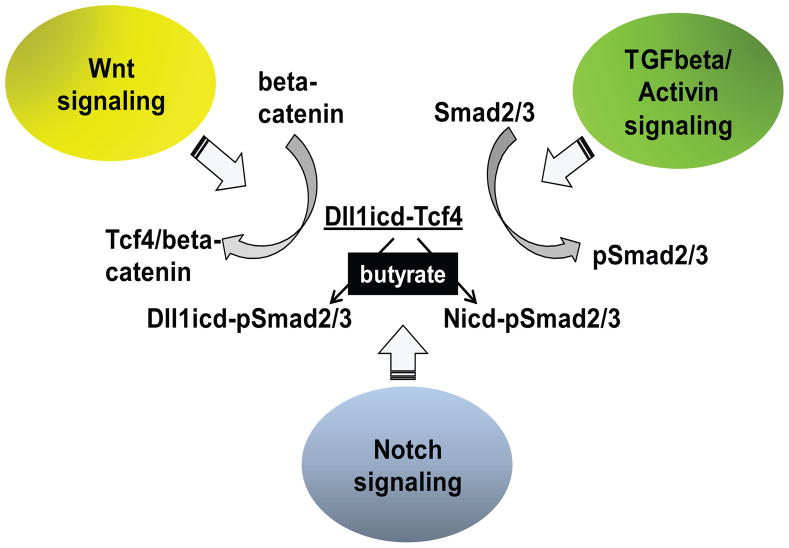
Based upon our findings, we propose a hypothetical model for the integration of three signaling pathways: Wnt, Notch, and TGFbeta/Activin. This integration may explain how colon cancer cell populations maintain heterogeneous levels of Wnt signaling (Fig.7). According to the model, a process similar to lateral inhibition translates the fluctuations in Notch and Dll1 levels among neighboring cells into different levels of Wnt activity in these cells. Thus, Smad2/3 proteins bind to the intracellular domains of Dll1 or Notch (Nicd or Dll1icd), dependent upon the availability of these molecules in the individual cells. The Dllicd1-Smad2/3 and Nicd-Smad2/3 complexes, which may form to different extent in adjacent cells of a cell population, act either as gene repressors or activators dependent upon the phosphorylation status of the Smad2/3 proteins. Cells, in which the Nicd-Smad2/3 complexes are at higher levels, suppress Wnt activity. The Nicd-Smad2/3 complexes may modulate gene expression through both CBF1 and Smad consensus binding elements [80–82]. Cells, in which the Dllicd1-Smad2/3 complexes are predominant, hyper-activate Wnt signaling in the presence of HDACis such as butyrate [13,18–20]. Since we have established that the fold induction of Wnt signaling correlates causatively with the levels of apoptosis in CC cells exposed to HDACis [13,18–20], understanding the mechanism for Wnt signaling heterogeneity in cancer cell populations may allow for the design of more effective anti-cancer approaches that eliminate malignant cells despite their different levels of Wnt signaling.
Fig. 7.
Integration of three signaling pathways. Wnt signaling results in formation of Tcf-4/beta- - catenin complexes which activate expression from Wnt targeted genes; TCFbeta/Activin signaling is mediated by phosphorylated Smad2/3. In mock-treated HCT-116 cells, a limited amount of Dll1icd is bound to Tcf4 species, and likely to dephosphorylated Smad2/3; these transcriptional complexes might be repressive. In butyrate-treated HCT-116 cells, the steady-state levels of the Tcf4 species decrease, and some of the Tcf4 protein pool is recruited to Wnt signaling complexes with dephosphorylated beta-catenin. Butyrate-treated cells also exhibit increased levels of phosphorylated Smad2/3 (pSmad2/3) proteins. In a fraction of the cell population, the pSmad2/3 proteins are bound to Dll1icd, and the resulting complexes may activate a specific set of genes (e.g., CTGF, p21, etc). In the remainder of the cell population, the pSmad2/3 proteins bind to Nicd, and these complexes may modulate a different set of genes [80–82]. Our preliminary data suggest that cells with higher levels of Dll1icd respond to butyrate with greater hyper-activation of Wnt signaling, and higher levels of apoptosis. The cells with higher levels of Nicd may suppress the hyper-activation of Wnt/beta-catenin by butyrate, and are likely more resistant to apoptosis.
Supplementary Material
Fig. 1S Specificity of anti-Dll1 antibody (Santa Cruz Biotechnology, cat. sc-8155) raised against a peptide mapping near the C-terminus of human Dll1. Protein lysates of HCT-116 cells that were exposed to mock (M) or 5 mM butyrate (B) treatment for 17h were analyzed via Western blotting. Two identical blots of the lysates were prepared and probed either with the sc-8155 antibody or with the same antibody pre-absorbed with a blocking peptide (sc-8155P).
Fig.2S The use of pGL3Promoter SV40 (pGL3) reporter instead of pGL3Basic (promoterless) reporter as the control in transcriptional analyses of the CTGF promoter activity confirms the induction of CTGF by butyrate, and the ability of dnTcf4 to suppress this induction. HCT-116 cells were co-transfected as described in Fig.4C: 80ng CTGF or pGL3Promoter (pGL3) luciferase reporter with 240ng dnTcf4 or pcDNA3, and pRLTK (at 1:100 ratio, to normalize for transfection efficiency). Cells were exposed to 5mM butyrate or mock treatment for 17h. The presence of dnTcf4 reduced the induction of CTGF/CTRL by butyrate from 3-fold to 1.5-fold (CTGF/CTRL is induced by butyrate from 4.2±0.82 to 12.8±1.1, P<0.001, in presence of pcDNA3, and from 4.2±0.46 to 6.2±1.1, P<0.05, in the presence of dnTcf4). Data are results from three separate experiments with duplicate samples for each treatment. Bars, SDs.
Acknowledgments
We thank Drs. I. Prudovsky, A. Leask, H. Clevers, D. Wu, B. Vogelstein, K. Kinzler, and D. Danielpour for sharing with us recombinant constructs. This work was supported by the NIH/NCI grant #1R15CA152852-01 and institutional funds.
Abbreviations
- ALK
Activin receptor-like kinases
- APC
adenomatous polyposis coli
- CC
colon cancer
- CTGF
connective tissue growth factor
- Dll1
Delta-like 1
- Dll1icd
Delta-like 1 intracellular domain
- HDACis
histone deacetylase inhibitors
- MEM
minimal essential media
- siRNA
small interfering RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Michael Bordonaro, Email: mbordonaro@tcmedc.org.
Shruti Tewari, Email: stewari@tcmedc.org.
Wafa Atamna, Email: watamna@tcmedc.org.
Darina L. Lazarova, Email: dlazarova@tcmedc.org.
References
- 1.Sancho E, Batlle E, Clevers H. Signaling pathways in intestinal development and cancer. Annu Rev Cell Dev Biol. 2004;20:695–723. doi: 10.1146/annurev.cellbio.20.010403.092805. [DOI] [PubMed] [Google Scholar]
- 2.Morin J, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 3.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell Biochem Biophys. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 4.Miyaki M, Iijima T, Kimura J, Yasuno M, Mori T, Hayashi Y, Koike M, Shitara N, Iwama T, Kuroki T. Frequent mutation of beta-catenin and APC genes in primary colorectal tumors from patients with hereditary nonpolyposis colorectal cancer. Cancer Res. 1999;59:4506–4509. [PubMed] [Google Scholar]
- 5.Roose J, Clevers H. Tcf transcription factors: molecular switches in carcinogenesis. Biochim Biophys Acta. 1999;87456:M23–M27. doi: 10.1016/s0304-419x(99)00026-8. [DOI] [PubMed] [Google Scholar]
- 6.Yochum GS, McWeeney S, Rajaraman V, Cleland R, Peters S, Goodman RH. Serial analysis of chromatin occupancy identifies beta-catenin target genes in colorectal carcinoma cells. Proc Natl Acad Sci USA. 2007;104:3324–3329. doi: 10.1073/pnas.0611576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatzis P, van der Flier LG, van Driel MA, Guryev V, Nielsen F, Denissov S, Nijman IJ, Koster J, Santo EE, Welboren W, Versteeg R, Cuppen E, van de Wetering M, Clevers H, Stunnenberg HG. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol Cell Biol. 2008;28:2732–2744. doi: 10.1128/MCB.02175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 9.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirements of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 10.Selkoe D, Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu Rev Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- 11.Schweisguth F. Notch signaling activity. Curr Biol. 2004;14:R129–R138. [PubMed] [Google Scholar]
- 12.Akiyama J, Okamoto R, Iwasaki M, Zheng X, Yui S, Tsuchiya K, Nakamura T, Watanabe M. Delta-like 1 expression promotes goblet cell differentiation in Notch-inactivated human colonic epithelial cells. Biochem Biophys Res Commun. 2010;393:662–667. doi: 10.1016/j.bbrc.2010.02.048. [DOI] [PubMed] [Google Scholar]
- 13.Lazarova DL, Bordonaro M, Sartorelli AC. Linear relationship between WNT activity -levels and apoptosis in colorectal carcinoma cells exposed to butyrate. Int J Cancer. 2004;110:523–531. doi: 10.1002/ijc.20152. [DOI] [PubMed] [Google Scholar]
- 14.Brabletz T, Jung A, Hermann K, Günther K, Hohenberger W, Kirchner T. Nuclear overexpression of the oncoprotein beta-catenin in colorectal cancer is localized predominantly at the invasion front. Pathol Res Pract. 1998;194:701–704. doi: 10.1016/s0344-0338(98)80129-5. [DOI] [PubMed] [Google Scholar]
- 15.Hlubek F, Brabletz T, Budczies J, Pfeiffer S, Jung A, Kirchner T. Heterogeneous expression of Wnt/beta-catenin target genes within colorectal cancer. Int J Cancer. 2007;121:1941–1948. doi: 10.1002/ijc.22916. [DOI] [PubMed] [Google Scholar]
- 16.Verras M, Papandreou I, Lim AL, Denko NC. Tumor hypoxia blocks Wnt processing and secretion through the induction of endoplasmic reticulum stress. Mol Cell Biol. 2008;28:7212–7224. doi: 10.1128/MCB.00947-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang D, Du X. Crosstalk between tumor cells and microenvironment via Wnt pathway in colorectal cancer dissemination. World J Gastroenterol. 2008;14:1823–1837. doi: 10.3748/wjg.14.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bordonaro M, Mariadason JM, Aslam F, Heerdt BG, Augenlicht LH. Butyrate-induced apoptotic cascade in colonic carcinoma cells: modulation of the beta-catenin-Tcf pathway and concordance with effects of sulindac and trichostatin A but not curcumin. Cell Growth Differ. 1999:713–720. [PubMed] [Google Scholar]
- 19.Bordonaro M, Lazarova DL, Augenlicht LH, Sartorelli AC. Cell type- and promoter-dependent modulation of the Wnt signaling pathway by sodium butyrate. Int J Cancer. 2002;97:42–51. doi: 10.1002/ijc.1577. [DOI] [PubMed] [Google Scholar]
- 20.Bordonaro M, Lazarova DL, Sartorelli AC. The activation of beta-catenin by Wnt signaling mediates the effects of histone deacetylase inhibitors. Exp Cell Res. 2007;313:1652–1666. doi: 10.1016/j.yexcr.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soler E, Andrieu-Soler C, Boer E, Bryne JC, Thongjuea S, Rijkers E, Demmers J, van Ijcken W, Grosveld F. A systems approach to analyze transcription factors in mammalian cells. 2010 doi: 10.1016/j.ymeth.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Lazarova DL, Bordonaro M, Sartorelli AC. Transcriptional regulation of the vitamin D (3) receptor gene by ZEB. Cell Growth Differ. 2001;12:319–326. [PubMed] [Google Scholar]
- 23.Bierie B, Moses HL. TGFbeta and cancer. Cytokine Growth Factor Rev. 2006;17:29–40. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Grady WM, Myeroff LL, Swinler SE, Rajput A, Thiagalingam S, Lutterbaugh JD, Neumann A, Brattain MG, Chang J, Kim SJ, Kinzler KW, Vogelstein B, Willson JK, Markowitz S. Mutational inactivation of transforming growth factor beta receptor type II in microsatellite stable colon cancers. Cancer Res. 1999;59:320–324. [PubMed] [Google Scholar]
- 25.Saif MW, Chu E. Biology of colorectal cancer. Cancer J. 2010;16:196–201. doi: 10.1097/PPO.0b013e3181e076af. [DOI] [PubMed] [Google Scholar]
- 26.Riggins GJ, Thiagalingam S, Rozenblum E, Weinstein CK, Kern SE, Hamilton SR, Willson JK, Markowitz SD, Kinzler KW, Vogelstein BB. Mad-related genes in the human. Nat Genet. 1996;13:347–349. doi: 10.1038/ng0796-347. [DOI] [PubMed] [Google Scholar]
- 27.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, Brattain M, Willson JKV. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 28.Thiagalingam S, Lengauer C, Leach FS, Schutte M, Hahn SA, Overhauser J, Willson JK, Markowitz S, Hamilton SR, Kern SE, Kinzler KW, Vogelstein B. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet. 1996;13:343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 29.Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui LC, Bapat B, Gallinger S, Andrulis IL, Thomsen GH, Wrana JL, Attisano L. MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell Biochem Biophys. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 30.Parsons R, Myeroff LL, Liu B, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- 31.Hempen PM, Zhang L, Bansal RK, Iacobuzio-Donahue CA, Murphy KM, Maitra A, Vogelstein B, Whitehead RH, Markowitz SD, Willson JK, Yeo CJ, Hruban RH, Kern SE. Evidence of selection for clones having genetic inactivation of the activin A type II receptor (ACVR2) gene in gastrointestinal cancers. Cancer Res. 2003;63:994–999. [PubMed] [Google Scholar]
- 32.Fink SP, Swinler SE, Lutterbaugh JD, Massagué J, Thiagalingam S, Kinzler KW, Vogelstein B, Willson JK, Markowitz S. Transforming growth factor-beta-induced growth inhibition in a Smad4 mutant colon adenoma cell line. Cancer Res. 2001;61:256–260. [PubMed] [Google Scholar]
- 33.Yanagisawa J, Yanagi Y, Masuhiro Y, Suzawa M, Watanabe M, Kashiwagi K, Toriyabe T, Kawabata M, Miyazono K, Kato S. Convergence of transforming growth factor-beta and vitamin D signaling pathways on SMAD transcriptional coactivators. Science. 1999;283:1317–1321. doi: 10.1126/science.283.5406.1317. [DOI] [PubMed] [Google Scholar]
- 34.Dai JL, Schutte M, Bansal RK, Wilentz RE, Sugar AY, Kern SE. Transforming growth factor-beta responsiveness in DPC4/SMAD4-null cancer cells. Mol Carcinog. 1999;26:37–43. doi: 10.1002/(sici)1098-2744(199909)26:1<37::aid-mc5>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 35.Sonoyama K, Pholnukulkit P, Toyoda M, Rutatip S, Kasai T. Upregulation of activin A gene by butyrate in human colon cancer cell lines. Am J Physiol Gastrointest Liver Physiol. 2003;284:G989–G995. doi: 10.1152/ajpgi.00384.2002. [DOI] [PubMed] [Google Scholar]
- 36.Pholnukulkit P, Sonoyama K, Kawabata J. Activin A induces phosphorylation of Smad2 but not complex formation of Smad2 with Smad4 in human colon cancer cell line HT-29. Biosci Biotechnol Biochem. 2003;67:2042–2044. doi: 10.1271/bbb.67.2042. [DOI] [PubMed] [Google Scholar]
- 37.Bahathiq AO, Stewart RL, Wells M, Moore HD, Pacey AA, Ledger WL. Production of activins by the human endosalpinx. J Clin Endocrinol Metab. 2002;87:5283–5289. doi: 10.1210/jc.2001-011884. [DOI] [PubMed] [Google Scholar]
- 38.Hiratochi M, Nagase H, Kuramochi Y, Koh CS, Ohkawara T, Nakayama K. The Delta intracellular domain mediates TGF-beta/Activin signaling through binding to Smads and has an important bi-directional function in the Notch-Delta signaling pathway. Nucleic Acids Res. 2007;35:912–922. doi: 10.1093/nar/gkl1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikeuchi T, Sisodia SS. The Notch ligands, Delta1 and Jagged2, are substrates for presenilin-dependent “gamma-secretase” cleavage. J Biol Chem. 2003;278:7751–7754. doi: 10.1074/jbc.C200711200. [DOI] [PubMed] [Google Scholar]
- 40.LaVoie MJ, Selkoe DJ. The Notch ligands, Jagged and Delta, are sequentially processed by alpha-secretase and presenilin/gamma-secretase and release signaling fragments. J Biol Chem. 2003;278:34427–34437. doi: 10.1074/jbc.M302659200. [DOI] [PubMed] [Google Scholar]
- 41.Six E, Ndiaye D, Laabi Y, Brou C, Gupta-Rossi N, Israel A, Logeat F. The Notch ligand Delta1 is sequentially cleaved by an ADAM protease and gamma secretase. Proc Natl Acad Sci USA. 2003;100:7638–7643. doi: 10.1073/pnas.1230693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee YJ, Swencki B, Shoichet S, Shivdasani RA. A possible role for the high mobility group box transcription factor Tcf-4 in vertebrate gut epithelial cell differentiation. J Biol Chem. 1999;274:1566–1572. doi: 10.1074/jbc.274.3.1566. [DOI] [PubMed] [Google Scholar]
- 43.Weise A, Bruser K, Elfet S, Wallmen B, Wittel Y, Wöhrle S, Hecht A. Alternative splicing of Tcf7l2 transcripts generates protein variants with differential promoter-binding and transcriptional activation properties at Wnt/beta-catenin targets. Nucl Acids Res. 2010;38:1964–1981. doi: 10.1093/nar/gkp1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galceran J, Sustmann C, Hsu SC, Folberth S, Grosschedl R. LEF1-mediated regulation of Delta-like1 links Wnt and Notch signaling in somitogenesis. Genes Dev. 2004;18:2718–2723. doi: 10.1101/gad.1249504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katoh M, Katoh M. Notch ligand, JAG1, is evolutionarily conserved target of canonical WNT signaling pathway in progenitor cells. Int J Mol Med. 2006;17:681–685. [PubMed] [Google Scholar]
- 46.Estrach S, Ambler CA, Lo Celso C, Hozumi K, Watt FM. Jagged 1 is a beta-catenin target gene required for ectopic hair follicle formation in adult epidermis. Dev. 2006;133:4427–4438. doi: 10.1242/dev.02644. [DOI] [PubMed] [Google Scholar]
- 47.Kolev V, Kacer D, Trifonova R, Small D, Duarte M, Soldi R, Graziani I, Sideleva O, Larman B, Maciag T, Prudovsky I. The intracellular domain of Notch ligand Delta1 induces cell growth arrest. FEBS Lett. 2005;579:5798–5802. doi: 10.1016/j.febslet.2005.09.042. [DOI] [PubMed] [Google Scholar]
- 48.Zarrinkalam KH, Stanley JM, Gray J, Oliver N, Faull RJ. Connective tissue growth factor and its regulation in the peritoneal cavity of peritoneal dialysis patients. Kidney Int. 2003;64:331–338. doi: 10.1046/j.1523-1755.2003.00069.x. [DOI] [PubMed] [Google Scholar]
- 49.Igarashi A, Okochi H, Bradham DM, Grotendorst GR. Regulation of connective tissue growth factor gene expression in human skin fibroblasts and during wound repair. Mol Biol Cell. 1993;4:637–645. doi: 10.1091/mbc.4.6.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abraham DJ, Shiwen X, Black CM, Sa S, Xu Y, Leask A. Tumor necrosis factor alpha suppresses the induction of connective tissue growth factor by transforming growth factor-beta in normal and scleroderma fibroblasts. J Biol Chem. 2000;275:15220–15225. doi: 10.1074/jbc.275.20.15220. [DOI] [PubMed] [Google Scholar]
- 51.Blom IE, Goldschmeding R, Leask A. Gene regulation of connective tissue growth factor: new targets for antifibrotic therapy? Matrix Biol. 2002;21:473–482. doi: 10.1016/s0945-053x(02)00055-0. [DOI] [PubMed] [Google Scholar]
- 52.Leask A. A sticky situation: CCN1 promotes both proliferation and apoptosis of cancer cells. J Cell Commun Signal. 2010;4:109–110. doi: 10.1007/s12079-009-0079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen S, McLean S, Carter DE, Leask A. The gene expression profile induced by Wnt 3a in NIH 3T3 fibroblasts. J Cell Commun Signal. 2007;1:175–183. doi: 10.1007/s12079-007-0015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen S, Leask A. Wnt 10b activates the CCN2 promoter in NIH 3T3 fibroblasts through the Smad response element. J Cell Commun Signal. 2009;3:57–59. doi: 10.1007/s12079-009-0049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H, Song K, Sponseller TL, Danielpour D. Novel Function of Androgen Receptor-associated Protein 55/Hic-5 as a Negative Regulator of Smad3 Signaling. J Biol Chem. 2005;280:5154–5162. doi: 10.1074/jbc.M411575200. [DOI] [PubMed] [Google Scholar]
- 56.Chipuk JE, Cornelius SC, Pultz NJ, Jorgensen JS, Bonham MJ, Kim SJ, Danielpour D. The Androgen Receptor Represses Transforming Growth Factor-beta Signaling through Interaction with Smad3. J Biol Chem. 2002;277:1240–1248. doi: 10.1074/jbc.M108855200. [DOI] [PubMed] [Google Scholar]
- 57.Bland CE, Kimberly P, Rand MD. Notch-induced proteolysis and nuclear localization of the Delta ligand. J Biol Chem. 2003;278:13607–13610. doi: 10.1074/jbc.C300016200. [DOI] [PubMed] [Google Scholar]
- 58.D'Souza B, Miyamoto A, Weinmaster GG. The many facets of Notch ligands. Oncogene. 2008;27:5148–5167. doi: 10.1038/onc.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kloth JN, Fleuren GJ, Oosting J, de Menezes RX, Eilers PHC, Kenter GG, Gorter A. Substantial changes in gene expression of Wnt, MAPK and TNFalpha pathways induced by TGF-beta1 in cervical cancer cell lines. Carcinogenesis. 2005;26:1493–1502. doi: 10.1093/carcin/bgi110. [DOI] [PubMed] [Google Scholar]
- 60.Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell Proteo. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 61.Datto MB, Yu Y, Wang XF. Functional analysis of the transforming growth factor beta responsive elements in the WAF1/Cip1/p21 promoter. J Biol Chem. 1995a;270:28623–28628. doi: 10.1074/jbc.270.48.28623. [DOI] [PubMed] [Google Scholar]
- 62.Barker N, Huls G, Korinek V, Clevers H. Restricted high level expression of Tcf-4 protein in intestinal and mammary gland epithelium. Am J Pathol. 1999;154:29–35. doi: 10.1016/S0002-9440(10)65247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gregorieff A, Pinto D, Begthel H, Destrée O, Kielman M, Clevers H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 64.Nawshad A, Medici D, Liu CC, Hay ED. TGFbeta3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad2-Smad4-LEF1 transcription complex. J Cell Sci. 2007;120:1646–1653. doi: 10.1242/jcs.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luo Q, Kang Q, Si W, Jiang W, Park JK, Peng Y, Li X, Luu HH, Luo J, Montag AG, Haydon RC, He TC. Connective tissue growth factor (CTGF) is regulated by Wnt and bone morphogenetic proteins signaling in osteoblast differentiation of mesenchymal stem cells. J Biol Chem. 2004;279:55958–55968. doi: 10.1074/jbc.M407810200. [DOI] [PubMed] [Google Scholar]
- 66.Zhang B, Zhou KK, Ma JX. Inhibition of CTGF Over-expression in Diabetic Retinopathy by SERPINA3K via Blocking the WNT/Beta-catenin Pathway. Diabetes. 2010;59:1809–1816. doi: 10.2337/db09-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Labbé E, Letamendia A, Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc Natl Acad Sci U S A. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hirota M, Watanabe K, Hamada S, Sun Y, Strizzi L, Mancino M, Nagaoka T, Gonzales M, Seno M, Bianco C, Salomon DS. Smad2 functions as a co-activator of canonical Wnt/beta-catenin signaling pathway independent of Smad4 through histone acetyltransferase activity of p300. Cell Signal. 2008;20:1632–1641. doi: 10.1016/j.cellsig.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang M, Wang M, Tang X, Li TF, Zhang Y, Chen DD. Smad3 prevents beta-catenin degradation and facilitates beta-catenin nuclear translocation in chondrocytes. J Biol Chem. 2010;285:8703–8710. doi: 10.1074/jbc.M109.093526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Labbé E, Lock L, Letamendia A, Gorska AE, Gryfe R, Gallinger S, Moses HL, Attisano L. Transcriptional cooperation between the transforming growth factor-beta and Wnt pathways in mammary and intestinal tumorigenesis. Cancer Res. 2007;67:75–84. doi: 10.1158/0008-5472.CAN-06-2559. [DOI] [PubMed] [Google Scholar]
- 71.Chu CY, Chang CC, Prakash E, Kuo ML. Connective tissue growth factor (CTGF) and cancer progression. J Biomed Sci. 2008;15:675–685. doi: 10.1007/s11373-008-9264-9. [DOI] [PubMed] [Google Scholar]
- 72.Yeger H, Perbal BB. The CCN family of genes: a perspective on CCN biology and therapeutic potential. J Cell Commun Signal. 2007;1:159–164. doi: 10.1007/s12079-008-0022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen PS, Wang MY, Wu SN, Su JL, Hong CC, Chuang SE, Chen MW, Hua KT, Wu YL, Cha ST, Babu MS, Chen CN, Lee PH, Chang KJ, Kuo ML. CTGF enhances the motility of breast cancer cells via an integrin-alphavbeta3-ERK1/2-dependent S100A4-upregulated pathway. J Cell Sci. 2007;120:2053–2065. doi: 10.1242/jcs.03460. [DOI] [PubMed] [Google Scholar]
- 74.Aikawa T, Gunn J, Spong SM, Klaus SJ, Korc M. Connective tissue growth factor-specific antibody attenuates tumor growth, metastasis, and angiogenesis in an orthotopic mouse model of pancreatic cancer. Mol Cancer Ther. 2006;5:1108–1116. doi: 10.1158/1535-7163.MCT-05-0516. [DOI] [PubMed] [Google Scholar]
- 75.Chang CC, Shih JY, Jeng YM, Su JL, Lin BZ, Chen ST, Chau YP, Yang PC, Kuo ML. Connective tissue growth factor and its role in lung adenocarcinoma invasion and metastasis. J Natl Cancer Inst. 2004;96:364–375. doi: 10.1093/jnci/djh059. [DOI] [PubMed] [Google Scholar]
- 76.Jiang WG, Watkins G, Fodstad O, Douglas-Jones A, Mokbel K, Mansel RE. Differential expression of the CCN family members Cyr61, CTGF and Nov in human breast cancer. Endo Relat Cancer. 2004;11:781–791. doi: 10.1677/erc.1.00825. [DOI] [PubMed] [Google Scholar]
- 77.Hishikawa K, Oemar BS, Tanner FC, Nakaki T, Lüscher TF, Fujii TT. Connective tissue growth factor induces apoptosis in human breast cancer cell line MCF-7. J Biol Chem. 1999;274:37461–37466. doi: 10.1074/jbc.274.52.37461. [DOI] [PubMed] [Google Scholar]
- 78.Ernst A, Campos B, Meier J, Devens F, Liesenberg F, Wolter M, Reifenberger G, Herold-Mende C, Lichter P, Radlwimmer B. De-repression of CTGF via the miR-17–92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene. 2010;29:3411–3422. doi: 10.1038/onc.2010.83. [DOI] [PubMed] [Google Scholar]
- 79.Hishikawa K, Oemar BS, Tanner FC, Nakaki T, Fujii T, Lüscher TF. Overexpression of connective tissue growth factor gene induces apoptosis in human aortic smooth muscle cells. Circulation. 1999;100:2108–2112. doi: 10.1161/01.cir.100.20.2108. [DOI] [PubMed] [Google Scholar]
- 80.Tang Y, Urs S, Boucher J, Bernaiche T, Venkatesh D, Spicer DB, Vary CP, Liaw L. Notch and transforming growth factor-beta (TGFbeta) signaling pathways cooperatively regulate vascular smooth muscle cell differentiation. J Biol Chem. 2010;285:17556–17563. doi: 10.1074/jbc.M109.076414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blokzjil A, Dahlqvist C, Reissman E, Falk A, Moliner A, Lendahl U, Ibanez CF. Cross-talk between the Notch and TGF-beta signaling pathways mediated by interaction of the Notch intracellular domain with Smad3. J Cell Biol. 2003;163:723–728. doi: 10.1083/jcb.200305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun Y, Lowther W, Kato K, Bianco C, Kenney N, Strizzi L, Raafat D, Hirota M, Khan NI, Bargo S, Jones B, Salomon D, Callahan R. Notch4 intracellular domain binding to Smad3 and inhibition of the TGF-beta signaling. Oncogene. 2005;24:5365–5374. doi: 10.1038/sj.onc.1208528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. 1S Specificity of anti-Dll1 antibody (Santa Cruz Biotechnology, cat. sc-8155) raised against a peptide mapping near the C-terminus of human Dll1. Protein lysates of HCT-116 cells that were exposed to mock (M) or 5 mM butyrate (B) treatment for 17h were analyzed via Western blotting. Two identical blots of the lysates were prepared and probed either with the sc-8155 antibody or with the same antibody pre-absorbed with a blocking peptide (sc-8155P).
Fig.2S The use of pGL3Promoter SV40 (pGL3) reporter instead of pGL3Basic (promoterless) reporter as the control in transcriptional analyses of the CTGF promoter activity confirms the induction of CTGF by butyrate, and the ability of dnTcf4 to suppress this induction. HCT-116 cells were co-transfected as described in Fig.4C: 80ng CTGF or pGL3Promoter (pGL3) luciferase reporter with 240ng dnTcf4 or pcDNA3, and pRLTK (at 1:100 ratio, to normalize for transfection efficiency). Cells were exposed to 5mM butyrate or mock treatment for 17h. The presence of dnTcf4 reduced the induction of CTGF/CTRL by butyrate from 3-fold to 1.5-fold (CTGF/CTRL is induced by butyrate from 4.2±0.82 to 12.8±1.1, P<0.001, in presence of pcDNA3, and from 4.2±0.46 to 6.2±1.1, P<0.05, in the presence of dnTcf4). Data are results from three separate experiments with duplicate samples for each treatment. Bars, SDs.