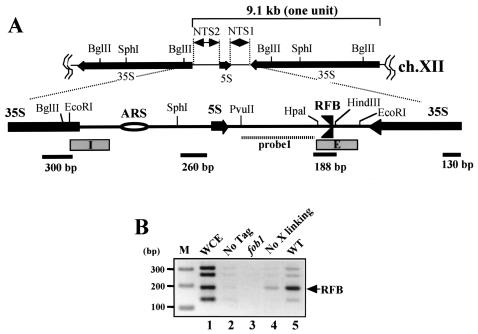
FIG. 1.
(A) Structure of rDNA repeats in S. cerevisiae. A single unit of rDNA consists of two transcribed genes (5S and 35S RNA genes) (the direction of transcription is indicated by arrows) and two nontranscribed regions (NTS1 and NTS2). The 35S rRNA gene is transcribed by PolI, while the 5S rRNA gene is transcribed by PolIII. The NTS and its surrounding regions are expanded. Two DNA elements related to DNA replication, the origin of replication (ARS) and the RFB, are located in NTS2 and NTS1, respectively. The RFB located near the end of the 35S rRNA gene allows progression of the replication fork in the direction of 35S rRNA transcription but not in the opposite direction (3, 35). Probe 1 (striped bar) is a probe used for Southern hybridization in 2D analysis. The lower bars show locations of fragments amplified by PCR for ChIP assay. The four fragments are located about every 1 kb. The lengths of the PCR products are shown below by bars. E and I (boxed) are elements of HOT1. (B) ChIP assay. The inverse image of PCR products resolved on ethidium bromide-stained 2.6% agarose gels is shown. Fob1-FLAGp was expressed in the fob1 strain (NOY408-1bf), and PCR was performed on chromatin fragments after the immunoprecipitation. Samples were prepared from the whole-cell extract (WCE) before the precipitation (lane 1), from NOY408-1b with Fob1p not tagged (lane 2), and from NOY408-1bf with Fob1-FLAGp not expressed (lane 3), with Fob1-FLAGp expressed but without prior cross-linking before precipitation (lane 4), and with Fob1-FLAGp expressed (lane 5). The position of the RFB fragment is indicated by an arrow. Lane M, 100-bp ladder marker (Invitrogen).