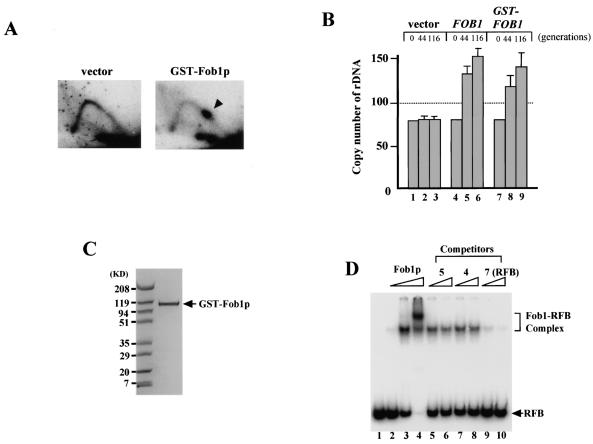
FIG. 4.
Binding activity of purified GST-Fob1p. (A) 2D analysis to detect replication fork-blocking activity of GST-Fob1p in vivo. DNA samples were prepared from NOY408-1bf carrying pEG(KT) (vector) or pTAK900 (GST-Fob1p). An arrowhead shows the accumulation of Y-shaped molecules, indicative of replication fork-blocking activity. (B) rDNA amplification activity of GST-Fob1p in vivo. NOY408-1af was transformed with pEG(KT) (vector), Yep-FOB1 (FOB1) or pTAK900 (GST-FOB1). At 44 and 116 generations after the introduction, DNA was isolated and rDNA copy number was determined. Generation 0 corresponds to DNA that was isolated before transformation. (C) Analysis of purified GST-Fob1p by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. GST-Fob1p was purified by using a GST-affinity column and gel filtration from a crude extract of YK9 with pTAK900. The fusion protein was applied to a 10 to 20% polyacrylamide gel and stained with Bio-Safe Coomassie blue (Bio-Rad). (D) Detection of in vitro binding activity of GST-Fob1p to the RFB fragment by gel shift assay. End-labeled RFB fragments (0.16 ng) (Fig. 3B, fragment 7) were mixed with 0 ng (lane 1) 0.25 ng (lane 2), 2.5 ng (lane 3), and 25 ng (lane 4) of GST-Fob1p, and the mixture was applied to a native 5% polyacrylamide gel. Lanes 5 to 10 show competition assays to detect the binding specificity of the RFB fragment. Here, 0.16 ng of end-labeled RFB fragments, 2.5 ng of GST-Fob1p, and one of the three kinds of cold competitor fragments (fragment 5, 4, or 7 [Fig. 3B]) were used in the assay. Fragments 5 and 4 are RFB flanking sequences, and fragment 7 is the RFB itself. Competitors were used at 1.6 ng (lanes 5, 7, and 9) or 3.2 ng (lanes 6, 8, and 10).