Abstract
Long under appreciated as important cellular organelles, lipid droplets are finally being recognized as dynamic structures with a complex and interesting biology. In light of this newfound respect, we discuss emerging views on lipid droplet biology and speculate on the major advances to come.
Long perceived as inert fat particles, lipid droplets (LDs) have been largely ignored by cell biologists. However, more recently, LDs are increasingly recognized as dynamic organelles that represent a frontier for cell biology.
These ubiquitous organelles are found in most eukaryotic cells (Figure 1; see SnapShot, this issue). They range greatly in size (diameter < 1–100 μm), and each consists of a phospholipid monolayer that surrounds a core of neutral lipids, such as sterol esters or triacylglycerols. Numerous proteins, many of which play functional roles in LD biology, decorate their surfaces. LDs are therefore structurally similar to plasma lipoproteins, which are secreted from cells and transport lipids through the aqueous circulation to different regions of the body.
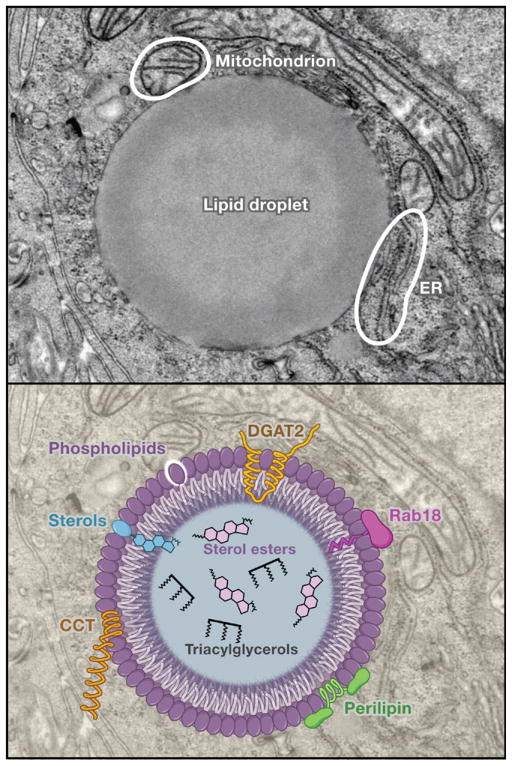
Figure 1. Anatomy of a Lipid Droplet.
(Top) An electron micrograph of a lipid droplet in a cultured hepatoma cell. The membrane monolayer surrounding the lipid droplet is visible, as are close associations with mitochondria and ER membranes. (Bottom) The structural features of a lipid droplet. Shown are polar surface lipids of the monolayer (e.g., phospholipids and sterols), the nonpolar lipids of the core (e.g., sterol esters and triacylglycerols), and a variety of proteins decorating the surface of the droplet. These proteins include DGAT2, Rab18, perilipin, and CCT (CTP:phosphocholine cytidylyltransferase; the rate-limiting enzyme in phosphatidylcholine synthesis). Several hypothetical mechanisms for how proteins interact with the lipid droplet are shown, including amphipathic α helices, embedding of hydrophobic regions directly in the droplet, and lipid anchors. (Electron micrograph courtesy of S. Stone and J. Wong; image reprinted from Biochim. Biophys. Acta 1791, T.C. Walter and R.V. Farese, Jr. (2009), with permission from Elsevier.)
We are just beginning to understand fundamental aspects of LD biogenesis, catabolism, and functional activities in cells. This increased knowledge of LD cell biology has relevance for applied science in the fields of human health and biofuels.
R—Recognition of Lipid Droplets as an Organelle
The earliest descriptions of lipid droplets date to the 19th century. Both Richard Altmann and E.B. Wilson described fat droplets in cells and speculated about their origin (Altmann, 1890; Wilson, 1896). Early on, the high diffraction properties of LDs facilitated their identification by light microscopy. With their recognition as a component of most cells in the early 1900s came a respectable name for this organelle: the liposome. However, in the late 1960s, artificial liposomes were invented and quickly usurped the name. Since then, the organelles were called by many names, including lipid droplets, lipid bodies, fat bodies, fat droplets, and adiposomes. In plants, they are often called oil bodies. As the field rapidly evolves, it seems to be settling on the name “lipid droplets.”
Other than morphological studies, LDs received scant attention for decades. In 1991, the discovery of perilipin, a phosphoprotein associated with LDs in adipocytes, brought new attention to the organelle (Greenberg et al., 1991). Since then, the number of papers about LDs has increased dramatically. This may reflect numerous factors, including basic research related to obesity and oil production, fields in which LD biology plays a prominent role, and the recognition of a virtually untapped frontier of cell biology.
E—Emerging Recognition of Functional Roles in Cells
For many cells and organisms, energy supplies in the environment fluctuate between surplus and starvation, and the ability to store energy may provide a competitive evolutionary advantage. Cellular energy is stored mainly in the form of triacylglycerols, which are hydrophobic, highly reduced, and concentrated molecules for storing energy. Within cells, energy storage is compartmentalized in LDs. Indeed, the most highly specialized cells dedicated to this process are adipocytes, in which LDs often occupy the bulk of the cytoplasm. In mammals, energy storage and catabolism in LDs are highly regulated by hormones and signaling pathways.
LDs are also a repository for the building blocks for biological membranes, such as phospholipids and sterols. When needed, these lipids can be generated from catabolism and mobilization of lipids in LDs. In yeast, such hydrolysis of triacylglycerols has been linked to the cell cycle and is coupled with rapid membrane expansion (Kurat et al., 2009).
By compartmentalizing lipids, LDs buffer cells from the toxic effects of excessive amounts of lipid. For example, macrophages can take up large amounts of cholesterol, which can trigger endoplasmic reticulum (ER) stress and eventually cell death by apoptosis or necrosis (Maxfield and Tabas, 2005). However, excess sterols can be detoxified by conversion to sterol esters and storage in LDs, thereby protecting the cells against toxicity. Similar buffering through esterification and partitioning into LDs may protect cells from other lipids or lipophilic substances that may be toxic in excess (e.g., fatty acids or retinol).
LDs are involved in intracellular protein metabolism. For example, during development of the fruit fly Drosophila, histones accumulate at the surfaces of LDs until they are incorporated into rapidly dividing nuclei (Cermelli et al., 2006). The core protein of the hepatitis C virus also localizes to LDs during viral replication in hepatocytes (Miyanari et al., 2007). LDs also have been linked functionally to the spliceosome and proteasome (Cho et al., 2007; Guo et al., 2008; Ohsaki et al., 2006). In the proteasome, LDs might serve as platforms for the deposition and degradation of some proteins.
S—Synthesis of Lipids for Growing Lipid Droplets
The growth of an LD requires the addition of both polar lipids at the droplet surface and neutral lipids within the hydrophobic core. The polar lipids include primarily phospholipids and sterols. Phospholipids may be derived from de novo synthesis or by conversion of glycerolipids, such as diacylglycerol derived from triacylglycerol hydrolysis. Although data are limited, the major phospholipids in LDs are phosphatidylcholine and phosphatidylethanolamine. The biosynthesis of these two phospholipids occurs primarily in the ER. It is unclear how a need for synthesis of phosphatidylcholine and phosphatidylethanolamine is sensed by a growing LD and how these phospholipids are added to the LD surface. In cells with limited phosphatidylcholine synthesis due to loss of CTP:phosphocholine cytidylyltransferase (the rate-limiting enzyme), LDs are larger than normal, possibly in part because there is limited phosphatidylcholine for LD surfaces thus promoting the fusion of LDs (Guo et al., 2008).
The growth of an LD can involve the addition of large amounts of triacylglycerols to its core, implying that triacylglycerol synthesis must be coordinated with LD growth. The synthesis of triacylglycerol in cells occurs mostly from the glycerol-phosphate pathway in which fatty acid moieties are added sequentially to a glycerol backbone. In the final step of triacylglycerol synthesis, diacylglycerol and fatty acyl CoAs are converted to triacylglycerol in a reaction catalyzed by acyl CoA:diacylglycerol acyltransferases (DGAT) (Yen et al., 2008). Many of the enzymes in the triacylglycerol synthetic pathway, including DGAT enzymes, are found in the ER and mitochondria or in mitochondria-associated membranes and occur in multiple isoforms (Coleman and Lee, 2004; Yen et al., 2008).
How newly synthesized triacylglycerol is delivered to the cores of nascent LDs is unclear. The prevailing model posits that triacylglycerol is synthesized within the ER and released between the leaflets of the bilayer membrane. When triacylglycerol accumulates over a certain threshold, a fat lens is believed to form in the bilayer and bulge into the cytoplasm through a budding process. How the triacylglycerol is channeled to budding domains, how budding occurs mechanistically, and how the directionality of the budding is achieved are not understood. Indeed, little experimental evidence supports the budding model, and alternative models have been proposed (reviewed in Walther and Farese, 2009). For example, a speculative model that we proposed is “vesicular budding,” in which newly formed lipids might fill the bilayer of existing membrane vesicles, effectively filling the vesicle with neutral lipids (Walther and Farese, 2009). In a variation of this model, the ER bilayer is filled with neutral lipids and the resulting bulge does not detach from the ER. In this scenario, LDs would constitute a specialized ER domain that remains connected to the ER bilayer. This would allow rapid exchange of lipids between the ER and LDs during synthesis and mobilization. However, when LDs grow, the high curvature of the membrane at the neck, formally resembling a hemifusion intermediate, would likely require stabilizing proteins.
The addition of triacylglycerol to the cores of LDs may also occur through the local synthesis of triacylglycerol at the surfaces of LDs or in ER membranes adjacent to droplets. Two recent studies demonstrated triacylglycerol synthesis in isolated droplets and linked this to the presence of enzymes (e.g., acyl-coenzyme A synthetase and DGAT2) that activate and esterify fatty acids (Fujimoto et al., 2007; Kuerschner et al., 2008).
Many LDs also contain large amounts of sterol esters. For example, sterol esters constitute about half of the neutral lipids in yeast LDs. In mammalian cells, sterol esters are the major neutral lipid in LDs of adrenocortical cells and macrophage foam cells in atherosclerotic lesions. Sterol esters are synthesized from sterols and fatty acyl CoA in reactions catalyzed by sterol-O-acyltransferases. These enzymes, like DGATs, are localized in the ER, and it is unclear how their products are channeled to LD cores.
LDs might also “grow” through fusion of smaller LDs into larger LDs, but this is likely to be a rare event in most cell types. Although a few studies show apparent fusion of LDs in time-lapse images (Boström et al., 2007; Guo et al., 2008), conclusive data for this mechanism are lacking. If fusion does occur, one possible mechanism is through simple phase coalescence. In fact, we speculate that specific proteins coating LD surfaces may prevent them from coalescing. This conjecture is supported by data in seeds of the model plant Arabidopsis thaliana in which deficiency of oleosin, the major structural LD protein, leads to apparent fusion of LDs (Siloto et al., 2006). Another possibility is that SNARE proteins mediate LD fusion (Boström et al., 2007), similar to their roles in the fusion of vesicle bilayers, although evidence supporting this conjecture is limited.
P—Protein Targeting to Lipid Droplets
The metabolic functions of LDs may be partially mediated by proteins bound to their surface. How proteins are specifically targeted to LDs is unknown. Molecular “zip codes,” such as sequences that direct proteins to the ER or mitochondria, have not been identified for LDs. Several distinct mechanisms might mediate targeting of proteins to LDs.
Similar to the binding of some apolipoproteins to lipoproteins, the binding of some proteins to LD surfaces involves amphipathic α helices. Perhaps the best studied example is Tip47, one of the family of LD-binding PAT proteins (perilipin, ADRP/adipophilin, Tip47, and related proteins). The crystal structure of a fragment of Tip47 revealed structural similarity to apolipoprotein (apo) E (Hickenbottom et al., 2004), with a four-helix bundle that contains amphipathic α helices. By analogy to apoE’s binding to lipoproteins, the four-helix bundle of Tip47 may open to facilitate the binding of the internal hydrophobic helices to the surface of LDs. Binding via amphipathic α helices is a likely mechanism for other proteins that are targeted to LD surfaces. How such proteins would distinguish between the LD surface monolayer and the bilayer of other organelle surfaces is unknown. One possibility is that LD specificity might be achieved by a dynamic interplay between specific surface lipids and the proteins that bind and modify them.
Proteins might also target LDs through a hydrophobic domain that embeds in the LD surface. This domain could be at the N or C terminus or lie within an internal region of a protein, which would result in surrounding hydrophilic regions of the protein projecting from the droplet surface. Examples of the latter include DGAT2, hepatitis C virus core protein, and caveolins (Boulant et al., 2006; Martin and Parton, 2006; Ostermeyer et al., 2004; Thiele and Spandl, 2008). Such a topology would enable proteins to move from a bilayer membrane to a monolayer LD surface. For example, caveolin is normally located in specific plasma membrane domains via a long hydrophobic stretch that is embedded in the membrane bilayer. When cells are incubated with fatty acids, a significant fraction of caveolin localizes to LDs (Robenek et al., 2004). A similar mechanism involving the ER bilayer and LDs has been suggested for DGAT2 (Stone et al., 2009; Thiele and Spandl, 2008). The targeting of proteins to LDs through lipid modifications, similar to mechanisms that target proteins to membrane bilayers, also seems likely, although there is no direct evidence as yet for this mechanism.
How such proteins would dynamically change their localization from a bilayer membrane to an LD monolayer is unclear. It seems unlikely that a long lipid-embedded domain is extracted from membranes during relocalization. If LDs remain contiguous with membranes, such as the ER, the protein might directly diffuse from the bilayer to the LD. Alternatively, membrane vesicular traffic may connect the plasma membrane or ER with LDs. Two lines of evidence support this possibility. First, biochemical approaches reconstituting in vitro vesicle formation from yeast microsomes have led to identification of a vesicle population that might connect the ER with LDs (Takeda and Nakano, 2008). Second, two independent RNAi screens identified a crucial function for the Arf1/COPI vesicular transport machinery in LD morphology and lipolysis (Beller et al., 2008; Guo et al., 2008). A defect in lipolysis in cells lacking Arf1/COPI components may be due at least partially to a requirement for trafficking of the adipose triglyceride lipase (ATGL), the major triglyceride lipase, to LDs (Beller et al., 2008; Soni et al., 2009). Given that Arf1/COPI proteins have a canonical function in vesicle generation in the Golgi apparatus, they might also function in vesicular trafficking to LDs. This model, however, presents a topological problem. How would vesicles surrounded by a bilayer membrane fuse with the monolayer surrounding the LDs? A membrane hemi-fusion of the outer vesicle bilayer with the surface monolayer of an LD would provide one solution for the transfer of lipids or proteins, but this idea is highly speculative.
Complicating the interpretation of most protein localization data is that some proteins observed in LDs may actually be localized to membranes close to the LDs. Many electron micrographs show ER membranes immediately adjacent to the LD surface, and light microscopy methods lack sufficient resolution to distinguish the exact localization of proteins between the LD surface and adjacent membranes. For example, DGAT2, an ER enzyme, localizes to LDs when cells are treated with fatty acids, but it is unclear if this reflects targeting to the LD surface or to an adjacent ER bilayer compartment (Kuerschner et al., 2008; Stone et al., 2009). Similarly, ADRP/adipophilin, which is considered a “classic” LD surface protein by light microscopy studies, has a significant pool in the LD-juxtaposed ER membrane (Robenek et al., 2006). Proteins that lack transmembrane domains but have hydrophobic domains could exchange between the two compartments, either by diffusion if the membranes were contiguous or by a shuttling mechanism.
E—Endoplasmic Reticulum: The Site of LD Biogenesis?
Although formal proof is lacking, the current model for LD biogenesis posits that the organelles are derived from the ER. This is logical inasmuch as the enzymes (e.g., DGAT and ACAT) that synthesize neutral lipids for the cores of LDs are localized primarily in the ER, and ER membranes are often found close to LDs (for examples, see Blanchette-Mackie et al., 1995). Additionally, a recent freeze-fracture study revealed that a large portion of LDs are surrounded by ER membranes, similar to an egg cup holding an egg (Robenek et al., 2006). In yeast, nascent LDs are almost universally found in close proximity with ER membranes (N. Krahmer, P. Mardones, and T.C.W., unpublished data; Goodman, 2008).
Contact sites between LDs and the ER could serve several functions. One can envision a model in which LDs bulging from the ER maintain a physical connection with the ER, which could be stabilized by a specialized set of proteins. Such a connection could facilitate the exchange of proteins or lipids between the compartments. During lipolysis, for example, the diacylglycerols and fatty acids generated could be directly transferred to the ER for phospholipid synthesis. Similarly, a sterol transfer protein that was identified on the surface of LDs could mediate the transport of released sterol back to the ER (Hynynen et al., 2009).
Additionally, LDs may associate functionally with other organelles. For example, in bacteria, which lack an ER, evidence suggests that LDs originate from the plasma membrane (Wältermann et al., 2005). Other studies have shown that LDs interact with membranes of peroxisomes, mitochondria, or replication vacuoles of intracellular parasites (reviewed in Goodman, 2008; Walther and Farese, 2009).
Few proteins have been identified as crucial for LD formation. Genomewide screens in Drosophila cells identified many genes whose loss of function results in fewer or smaller lipid droplets (Beller et al., 2008; Guo et al., 2008). Similar screens in yeast suggest that seipin (encoded by the FLD1 gene) is involved (Fei et al., 2008; Szymanski et al., 2007), but mechanistic insights are lacking. Tip47 also has been implicated in LD biogenesis (Bulankina et al., 2009).
C—Catabolism of Lipid Droplets
When cells need lipids to generate energy or synthesize membranes, lipolysis is activated to mobilize these substrates. During lipolysis of triacylglycerols, individual fatty acyl chains are sequentially cleaved from the glycerol backbone. In mammalian adipose tissue, lipolysis is highly regulated and induced by catecholamines during fasting or exercise (Zechner et al., 2009). Catecholamines bind to G protein-coupled receptors, leading to an increase in intracellular cAMP via activation of adenylate cyclase and, as a consequence, the activation of protein kinase A (PKA). In turn, PKA phosphorylates a number of proteins, including hormone-sensitive lipase and perilipin (Brasaemle et al., 2009). Phosphorylation of perilipin triggers release of CGI-58, a coactivator of ATGL. Subsequently, ATGL is recruited to LDs, where the complex catalyzes the removal of the first acyl chain from triacylglycerols. Phosphorylation of hormone-sensitive lipase activates this enzyme, leading to relocalization to the LD surface, where it cleaves the second acyl chain, to yield monoacylglycerol. The last acyl chain is removed by monoacylglycerol lipase, and glycerol is released. During this process, the different lipases are targeted directly to the surface of the LD to interact with their respective substrates. But how, for example, ATGL penetrates the LD-delimiting monolayer and accesses triacylglycerols located in the core is unknown. We speculate that a lipase, or a protein complex involving a lipase, binds to the LD surface and effectively parts the surface phospholipid monolayer, allowing a neutral lipid in the core to access the catalytic site of an overlying lipase. Some evidence from cultured cells suggests that, under maximum stimulation of lipolysis, LDs may break up into smaller LDs, which would provide more surface area for lipases. Whether this occurs in normal physiology is unknown. Such a process could be facilitated by proteins that deform membranes and initiate budding, such as coatamer proteins (Guo et al., 2008). LDs may also get degraded by autophagy (Singh et al., 2009), but the relative contribution of this pathway to lipolysis is not clear. Autophagy could provide a mechanism for bulk recycling of lipids for cellular utilization, in contrast to the more selective, and hormonally induced, lipolytic mechanisms that provide release of lipids from cells.
T—Translational Relevance and Tomorrow’s Research
LDs play an important role in diseases of excess triacylglycerol storage, such as obesity and metabolic syndrome, which often lead to cardiovascular complications and the development of type 2 diabetes. Indeed, many diseases, such as obesity, diabetes, and fatty liver disease, could be referred to as “diseases of excessive LDs.” LDs in adipocytes, muscle cells, hepatocytes, and cardiac myocytes likely protect cells from the effects of too much fat. Yet, in many instances, excessive lipid deposition may exceed the capacity of cells and lead to dysfunction, termed lipotoxicity (reviewed in Schaffer, 2003). This raises the interesting and so far unanswered question of what determines the capacity of a cell to store fat. Several studies have shown that increasing the capacity for triacylglycerol synthesis in cells or tissues is protective in some instances (Yen et al., 2008). Interestingly, the protein Fsp27/CIDEC regulates fat storage in adipocytes (Gong et al., 2009) and might also regulate the storage capacity of LDs (Keller et al., 2008). Also, rare genetic disorders result in excessive LDs in tissues: homozygous deficiency of ATGL or CGI-58 causes neutral lipid storage disease and is associated with myopathy or skin ichthyosis, respectively (Schweiger et al., 2009).
LDs also play a major role in atherosclerosis. In atherosclerotic arteries, cholesterol esters accumulate in LDs of macrophages resulting in “foam cells,” which are hallmarks of atherosclerotic lesions. In foam cells, LDs likely serve to buffer the cells against toxicity from excessive amounts of unesterified sterols. If macrophages are overwhelmed by excessive amounts of lipids, they may undergo apoptotic and necrotic cell death, contributing to lesion instability and, ultimately, to stroke and heart attack (Maxfield and Tabas, 2005). Besides metabolic diseases, LDs function in the intracellular replication of several infectious agents, including Chlamydia and hepatitis C virus (Boulant et al., 2007; Cocchiaro et al., 2008; Ogawa et al., 2009), suggesting new therapeutic angles for these infections.
Apart from diseases, LDs are engendering intense study in the applied science of oil production for nutrition or biofuel purposes. Generating animals or crops with altered or maximized lipid production has long occurred through breeding and will likely increase with genetic engineering. Additionally, efforts to industrially produce LDs in algae or bacteria as sources of biofuels to provide energy are rapidly increasing (Service, 2009). Traditionally, plant oils, stored in lipid bodies (the functional equivalents of LDs in plants), were produced in species such as Brassica napus and processed to yield biodiesel. The current engineering of photosynthetic microorganisms such as algae promises to bring industrial scale production of oil to a new arena.
Conclusion
We have touched on most of the fundamental questions concerning LDs that remain unanswered, such as those pertaining to their functions, formation, growth, and catabolism. One additional question is whether all organelles now called LDs are indeed equivalent or merely resemble each other. For example, are the sterol-rich droplets in macrophage foam cells similar in protein composition to lipid droplets in triacylglycerol-rich adipocytes? Might different types of LDs originate through different mechanisms? At least for LDs formed in bacteria, it is already clear that the mechanisms differ from those in eukaryotes.
These questions provide a wealth of research opportunities, from biophysics to cell biology to lipid and energy metabolism in multicellular organisms. Some challenges that must be solved are methodological, and advances in these areas would greatly serve the field. For example, the application of high-resolution techniques of light microscopy might provide insights into the relationship of LDs with other organelles, such as the ER. Biochemical techniques to purify subpopulations of LDs, analogous to techniques to separate classes of plasma lipoproteins, might be extremely valuable. The development of methods to generate artificial LDs, similar to bilayer liposomes, would enable detailed biochemical and biophysical studies of phenomena such as LD fusion or protein targeting.
The field of LD biology is on the cusp of a new era of information. LDs are emerging from relative obscurity to recognition as dynamic structures with a complex and interesting biology that is highly integrated with other aspects of cell biology. Insights into the molecular aspects of LD biology will provide new information about physiology and disease and will provide fresh leads for the applied sciences. LDs are finally garnering the respect they richly deserve.
Acknowledgments
We thank members of the Farese and Walther laboratories, and in particular E. Currie, N. Krahmer, and C. Harris, for helpful discussions, and G. Howard for editorial assistance. This work was supported by NIH grant DK-056084 and the J. David Gladstone Institutes (to R.V.F.) and the Max Planck Society, the German Research Council (DFG), and the Human Frontier Science Program (to T.C.W.).
References
- Altmann R. Die Elementarorganisem und ihre Beziehungen zu den Zellen. Leipzig: Veit; 1890. [Google Scholar]
- Beller M, Sztalryd C, Southall N, Bell M, Jäckle H, Auld D, Oliver B. PLoS Biol. 2008;6:e292. doi: 10.1371/journal.pbio.0060292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette-Mackie EJ, Dwyer NK, Barber T, Coxey RA, Takeda T, Rondinone CM, Theodorakis JL, Greenberg AS, Londos C. J Lipid Res. 1995;36:1211–1226. [PubMed] [Google Scholar]
- Boström P, Andersson L, Rutberg M, Perman J, Lidberg U, Johansson B, Fernandez-Rodriguez J, Ericson J, Nilsson T, Borén J, et al. Nat Cell Biol. 2007;9:1286–1293. doi: 10.1038/ncb1648. [DOI] [PubMed] [Google Scholar]
- Boulant S, Montserret R, Hope R, Ratinier M, Targett-Adams P, Lavergne J, Penin F, McLauchlan J. J Biol Chem. 2006;281:22236–22247. doi: 10.1074/jbc.M601031200. [DOI] [PubMed] [Google Scholar]
- Boulant S, Targett-Adams P, McLauchlan J. J Gen Virol. 2007;88:2204–2213. doi: 10.1099/vir.0.82898-0. [DOI] [PubMed] [Google Scholar]
- Brasaemle D, Subramanian V, Garcia A, Marcinkiewicz A, Rothenberg A. Mol Cell Biochem. 2009;326:15–21. doi: 10.1007/s11010-008-9998-8. [DOI] [PubMed] [Google Scholar]
- Bulankina A, Deggerich A, Wenzel D, Mutenda K, Wittmann J, Rudolph M, Burger K, Höning S. J Cell Biol. 2009;185:641–655. doi: 10.1083/jcb.200812042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermelli S, Guo Y, Gross S, Welte M. Curr Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- Cho SY, Shin ES, Park PJ, Shin DW, Chang HK, Kim D, Lee HH, Lee JH, Kim SH, Song MJ, et al. J Biol Chem. 2007;282:2456–2465. doi: 10.1074/jbc.M608042200. [DOI] [PubMed] [Google Scholar]
- Cocchiaro J, Kumar Y, Fischer E, Hackstadt T, Valdivia R. Proc Natl Acad Sci USA. 2008;105:9379–9384. doi: 10.1073/pnas.0712241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RA, Lee DP. Prog Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- Fei W, Shui G, Gaeta B, Du X, Kuerschner L, Li P, Brown A, Wenk M, Parton R, Yang H. J Cell Biol. 2008;180:473–482. doi: 10.1083/jcb.200711136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto Y, Itabe H, Kinoshita T, Homma K, Onoduka J, Mori M, Yamaguchi S, Makita M, Higashi Y, Yamashita A, et al. J Lipid Res. 2007;48:1280–1292. doi: 10.1194/jlr.M700050-JLR200. [DOI] [PubMed] [Google Scholar]
- Gong J, Sun Z, Li P. Curr Opin Lipidol. 2009;20:121–126. doi: 10.1097/MOL.0b013e328328d0bb. [DOI] [PubMed] [Google Scholar]
- Goodman JM. J Biol Chem. 2008;283:28005–28009. doi: 10.1074/jbc.R800042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg A, Egan JJ, Wek S, Garty N, Blanchette-Mackie E, Londos C. J Biol Chem. 1991;266:11341–11346. [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV., Jr Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickenbottom S, Kimmel A, Londos C, Hurley J. Structure. 2004;12:1199–1207. doi: 10.1016/j.str.2004.04.021. [DOI] [PubMed] [Google Scholar]
- Hynynen R, Suchanek M, Spandl J, Bäck N, Thiele C, Olkkonen V. J Lipid Res. 2009;50:1305–1315. doi: 10.1194/jlr.M800661-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P, Petrie J, De Rose P, Gerin I, Wright WS, Chiang SH, Nielsen AR, Fischer CP, Pedersen BK, MacDougald OA. J Biol Chem. 2008;283:14355–14365. doi: 10.1074/jbc.M708323200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuerschner L, Moessinger C, Thiele C. Traffic. 2008;9:338–352. doi: 10.1111/j.1600-0854.2007.00689.x. [DOI] [PubMed] [Google Scholar]
- Kurat C, Wolinski H, Petschnigg J, Kaluarachchi S, Andrews B, Natter K, Kohlwein S. Mol Cell. 2009;33:53–63. doi: 10.1016/j.molcel.2008.12.019. [DOI] [PubMed] [Google Scholar]
- Martin S, Parton RG. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- Maxfield FR, Tabas I. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- Ogawa K, Hishiki T, Shimizu Y, Funami K, Sugiyama K, Miyanari Y, Shimotohno K. Proc Jpn Acad, Ser B, Phys Biol Sci. 2009;85:217. doi: 10.2183/pjab.85.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T. Mol Biol Cell. 2006;17:2674–2683. doi: 10.1091/mbc.E05-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeyer A, Ramcharan L, Zeng Y, Lublin D, Brown D. J Cell Biol. 2004;164:69–78. doi: 10.1083/jcb.200303037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robenek H, Hofnagel O, Buers I, Robenek MJ, Troyer D, Severs NJ. J Cell Sci. 2006;119:4215–4224. doi: 10.1242/jcs.03191. [DOI] [PubMed] [Google Scholar]
- Robenek MJ, Severs NJ, Schlattmann K, Plenz G, Zimmer KP, Troyer D, Robenek H. FASEB J. 2004;18:866–868. doi: 10.1096/fj.03-0782fje. [DOI] [PubMed] [Google Scholar]
- Schaffer JE. Curr Opin Lipidol. 2003;14:281–287. doi: 10.1097/00041433-200306000-00008. [DOI] [PubMed] [Google Scholar]
- Schweiger M, Lass A, Zimmerman R, Eichmann T, Zechner R. Am J Physiol Endocrinol Metab. 2009;297:E289–E296. doi: 10.1152/ajpendo.00099.2009. [DOI] [PubMed] [Google Scholar]
- Service R. Science. 2009;325:379. doi: 10.1126/science.325_379a. [DOI] [PubMed] [Google Scholar]
- Siloto R, Findlay K, Lopez-Villalobos A, Yeung E, Nykiforuk C, Moloney M. Plant Cell. 2006;18:1961–1974. doi: 10.1105/tpc.106.041269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo A, Czaja M. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni K, Mardones G, Sougrat R, Smirnova E, Jackson C, Bonifacino J. J Cell Sci. 2009;122:1834–1841. doi: 10.1242/jcs.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S, Levin M, Zhou P, Han J, Walther T, Farese RV., Jr J Biol Chem. 2009;284:5352–5361. doi: 10.1074/jbc.M805768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski K, Binns D, Bartz R, Grishin N, Li W, Agarwal A, Garg A, Anderson R, Goodman J. Proc Natl Acad Sci USA. 2007;104:20890–20895. doi: 10.1073/pnas.0704154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y, Nakano A. J Biochem (Tokyo) 2008;143:803–811. doi: 10.1093/jb/mvn034. [DOI] [PubMed] [Google Scholar]
- Thiele C, Spandl J. Curr Opin Cell Biol. 2008;20:378–385. doi: 10.1016/j.ceb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Wältermann M, Hinz A, Robenek H, Troyer D, Reichelt R, Malkus U, Galla HJ, Kalscheuer R, Stöveken T, von Landenberg P, et al. Mol Microbiol. 2005;55:750–763. doi: 10.1111/j.1365-2958.2004.04441.x. [DOI] [PubMed] [Google Scholar]
- Walther T, Farese RV., Jr Biochim Biophys Acta. 2009;1791:459. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E. The Cell in Development and Inheritance. New York: Macmillan; 1896. [Google Scholar]
- Yen C, Stone S, Koliwad S, Harris C, Farese R. J Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R, Kienesberger P, Haemmerle G, Zimmermann R, Lass A. J Lipid Res. 2009;50:3–21. doi: 10.1194/jlr.R800031-JLR200. [DOI] [PubMed] [Google Scholar]