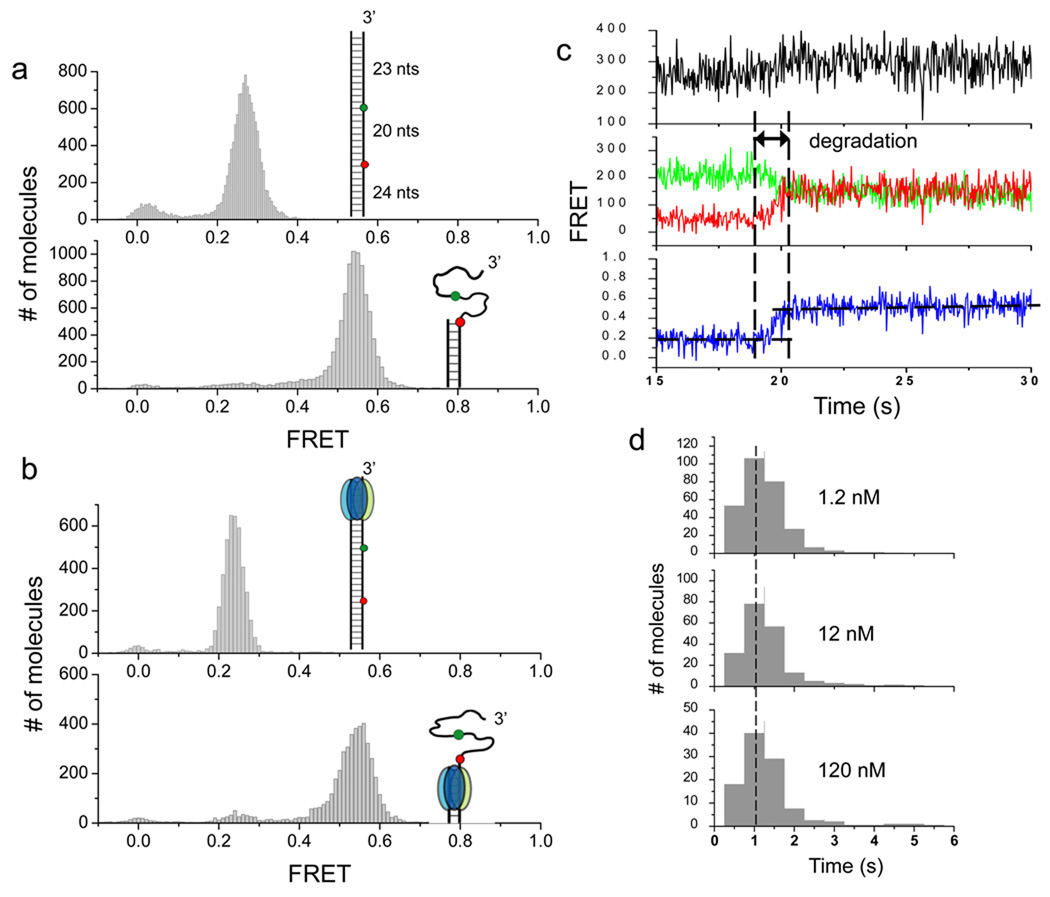
Figure 3.
After stable engagement with the 3’ overhang, λ exo processively degrades the substrate without dissociating. (a) In order to detect DNA degradation after the protein has been stabled engaged with the substrate, the original DNA construct used in Figures 1 and 2 are extended by 23 bp to its upstream (inset of the upper panel). The extension of 23 bp is longer than the footprint of the enzyme (13–14 nt), ensuring that the substrate can be fully engaged when the enzyme passes the region between two fluorophores (inset, upper panel). A partial duplex mimicking the reaction product was also constructed (inset, lower panel). Single molecule FRET histograms of the two constructs in the absence of the protein are shown. (b) Single molecule FRET histograms obtained after λ exo was injected into reaction chambers without Mg2+ (upper panel) and with Mg2+ (lower panel). Comparison of histograms between a and b verified that DNA degradation indeed occurred. (c) A representative set of time traces from a single molecule of the extended construct during degradation (120 nM protein). The degradation period measured by FRET is marked. (d) Histograms of degradation times at different protein concentrations are identical to each other, showing that the FRET increase from the extended construct reports on the processive phase.