Fig. 1.
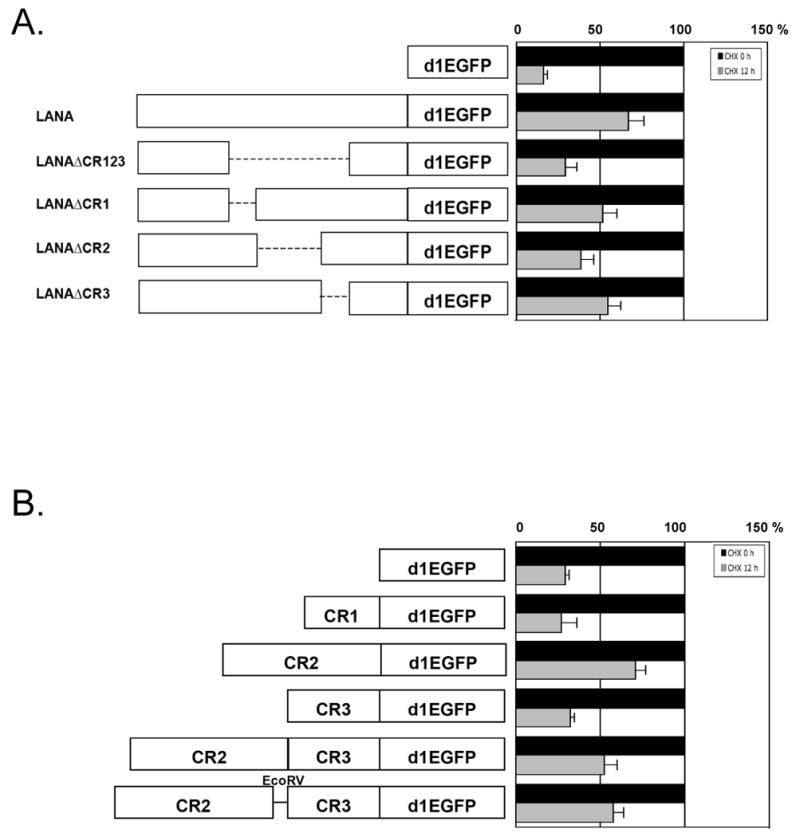
The central repeat (CR) 2 domain of LANA1 inhibits LANA1 turnover. (A) LANA1 repeat regions (CR1, CR2, CR3) were individually and in combination deleted from full-length LANA1, and cloned as fusion proteins with a destabilized enhanced green fluorescent protein (d1EGFP) (left panel) to ensure rapid proteasomal turnover. P value=0.007 (d1EGFP and LANAd1EGFP), 0.065 (LANAd1EGFP and LANAΔCR123d1EGFPd), 0.0225 (LANAd1EGFP and LANAΔCR2d1EGFP). (B) Turnover of individual LANA1 CR regions and subregions cloned into d1EGFP were also determined. P value=0.0441 (d1EGFP and CR2-d1EGFP), 0.022 (CR2-d1EGFP and CR1-d1EGFP), 0.0106 (CR2-d1EGFP and CR3-d1EGFP), 0.0325 (CR2-d1EGFP and CR2CR3-d1EGFP). Normalized EGFP fluorescence at time 0 (A and B right panel, black bars) was used as a reference to compare EGFP fluorescence 12 hours after CHX treatment (grey bars) to assess protein turnover. Results are average values performed in triplicate from independent experiments. P values between different groups, were obtained by two-tailed Student's t test using Prism software (GraphPad).
