Abstract
Transcription by RNA polymerase I in Saccharomyces cerevisiae requires a series of transcription factors that have been genetically and biochemically identified. In particular, the core factor (CF) and the upstream activation factor (UAF) have been shown in vitro to bind the core element and the upstream promoter element, respectively. We have analyzed in vivo the DNAse I footprinting of the 35S promoter in wild-type and mutant strains lacking one specific transcription factor at the time. In this way we were able to unambiguously attribute the protections by the CF and the UAF to their respective putative binding sites. In addition, we have found that in vivo a binding hierarchy exists, the UAF being necessary for CF binding. Because the CF footprinting is lost in mutants lacking a functional RNA polymerase I, we also conclude that the final step of preinitiation-complex assembly affects binding of the CF, stabilizing its contact with DNA. Thus, in vivo, the CF is recruited to the core element by the UAF and stabilized on DNA by the presence of a functional RNA polymerase I.
INTRODUCTION
In the yeast Saccharomyces cerevisiae, the information for the rRNA is stored in a locus on chromosome XII, the rDNA (Petes, 1979). The 35S gene localizes on the ribosomal locus as a tandem array of 150–200 copies, each one separated by the Non Transcribed Spacer (NTS). Previous studies indicate that cells adjust the rate of rRNA transcription according to their requirement for protein synthesis (Waldron and Lacroute, 1975; Kief and Warner, 1981).
Several cis-acting elements and trans-acting factors are known to coordinately assist transcription of rDNA by the RNA polymerase I. Moreover, a series of ancillary elements and factors have also been reported, but their ultimate roles in RNA polymerase I regulation have not been clarified yet.
The major DNA sequence elements involved in rDNA transcription are: the core element (CE; Musters et al., 1989; Kulkens et al., 1991; Keener et al., 1998), the upstream promoter element (UPE), and the enhancer element (Elion and Warner, 1986; Morrow et al., 1993; Schultz et al., 1993).
In a genetic screen for mutants that affect RNA polymerase I transcription, Nomura and collaborators isolated rrn mutants in yeast (Nogi et al., 1991). rrn mutants were found to be affected in the RNA polymerase I subunits or in RNA polymerase I transcription factors. Extracts from rrn mutants, identified as RNA polymerase I transcription factors, were used for in vitro transcription assays on rDNA templates (Keys et al., 1994). Purified factors can complement the mutant extracts, allowing transcription. This experimental approach showed that 1) the products of the RRN6, RNN7, and RRN11 genes form the core factor (CF), a complex that binds the CE and that is absolutely required for RNA polymerase I transcription (Keys et al., 1994; Lalo et al., 1996; Lin et al., 1996); 2) the RRN5, RRN9, and RRN10 gene products form the upstream activation factor (UAF) that binds the UPE; this factor is not essential, but it is stimulatory for 35S rRNA transcription and is directly involved in the recruitment of the CF to the promoter by interacting with the TATA-binding protein (TBP; Steffan et al., 1996; Steffan et al., 1998); 3) the RRN3 gene product represents an essential factor for the RNA polymerase I (Yamamoto et al., 1996) and acts by directly binding the RNA polymerase I. The exact role of Rrn3p has not been established, although it may be involved in the recruitment of RNA polymerase I or take part in the elongation process (Nomura, 1998; Reeder, 1999).
The factors described above were found in a genetic screen (Nogi et al., 1991) for mutants that abolish RNA polymerase I transcription or reduce it to a very low level. However, other factors are required for rDNA transcription, although their presence is not essential. Reb1p, a factor involved in the termination of transcription, recognizes a specific DNA-binding site, two copies of which are present in the NTS of rDNA (Reeder, 1999). We have recently reported the in vivo DNA footprint of Reb1p on the promoter (Vogelauer et al., 1998). DNA topoisomerase I is also most likely implicated in rDNA transcription. Its presence in vivo in the NTS was assessed by in vivo DNAse I footprinting (Vogelauer et al., 1998) and by the detection of its cleavage sites using the DNA topoisomerase I inhibitor camptothecin (Vogelauer and Camilloni, 1999).
In spite of the great detail of knowledge about RNA polymerase I transcription and regulation, a direct inspection of DNA-protein interactions occurring in vivo is still missing. By using the in vivo footprinting technique, we intend to show the binding sites for defined transcription factors in strains lacking the CF and UAF functions to unambiguously assign the footprints of these factors; in addition, we want to analyze possible interferences among specific factors, which can clarify the reciprocal roles of all factors in binding and assembling on the promoter to efficiently assist the RNA polymerase I.
MATERIALS AND METHODS
Yeast Strains, Plasmids, and Culture Media
The strains used in this study were NOY505 (Mata, ade 2-1, ura 3-1, his 3-11, trp1-1, leu2-3, 112, can1-100), NOY699 (Mata, ade 2-1, ura 3-1, his 3-11, trp1-1, leu2-3, 112, can1-100 rrn5::LEU2) pNOY103, NOY558 (Mata, ade 2-1, ura 3-1, his 3-11, trp1-1, leu2-3, 112, can1-100 rrn7::LEU2) pNOY103, and NOY604 (Mata, ade 2-1, ura 3-1, his 3-11, trp1-1, leu2-3, 112, can1-100 rrn3Δ::HIS3) pNOY103, which were kindly provided by M. Nomura. D128–1d (Mata, rpa43::LEU2 ade 2-101 uaa, ura 3-52, lys2-801 uag, trp1-Δ63, his 3-Δ200, leu 2-Δ1/) pNOY102 was kindly provided by P. Thuriaux. All the strains were grown in complete YPGal medium (Sherman et al., 1983) with 3% galactose.
Enzymes and Chemicals
DNAse I and T4 polynucleotide kinase were purchased from Roche (Indianapolis, IN), Vent (exo−) polymerase was from New England Biolabs (Beverly, MA), Zymolyase 100T was from Seikagaku (Tokyo, Japan), and radiochemicals were from Amersham (Arlington Heights, IL).
Preparation of Nuclei
Cells (150 ml grown to 0.4 OD600/ml) were centrifuged and resupended in 10 ml of a buffer containing 1 M sorbitol, 50 mM Tris-HCl, pH 7.5, and 10 mM β-mercaptoethanol, in the presence of 0.05 mg/3 × 107 cells of Zymolyase 100T, and incubated for 10 min at 30°C. Spheroplasts were then washed once with 1 M sorbitol and resuspended in lysis buffer (18% Ficoll, 20 mM potassium phosphate buffer, pH 6.8, 250 μM EDTA, 250 μM EGTA, 1 μM leupeptine, 1 mM phenylmethylsulfonyl fluoride, 0.15 mM spermine, 0.5 mM spermidine). Nuclei were prepared according to the method of Almer and Horz (1986), with minor modifications.
DNAse I Treatment
Nuclei were resuspended in digestion buffer (15 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1.4 mM CaCl2, 200 μM EDTA, 200 μM EGTA, 1 μM leupeptine, 1 mM phenylmethylsulfonyl fluoride, 0.15 mM spermine, 0.5 mM spermidine, 5 mM β-mercaptoethanol) and divided into 0.2-ml aliquots. DNAse I (3, 6, 12, 24 U) was added to each aliquot and the incubation was carried out at 0°C for 5 min. The reaction was stopped with 1% SDS and 5 mM EDTA (final concentrations). Proteinase K (40 μg/sample) was added, and the samples were kept at 56°C for 2 h. The DNA was then purified by three phenol/chloroform extractions and ethanol precipitation, followed by RNase A treatment.
Primers
The r3 synthetic oligonucleotide used as primer in the extension reactions lies at positions −267/−248 base pairs (bp; r3) from the RNA initiation site (RIS) (sequence number + 1; see also Figure 1). 5′-End labeling using [32P]γ-ATP and T4 polynucleotide kinase was performed according to standard procedures (Sambrook et al., 1989). The labeled oligonucleotides were purified by PAGE.
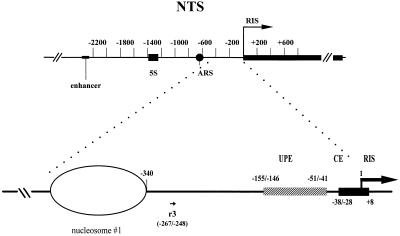
Figure 1.
Schematic representation of 35S RNA promoter in S. cerevisiae. Numbering is relative to the 35S transcriptional start (RIS). The ellipse indicates the first of five phased nucleosomes lying in the NTS. Positions of the most important DNA element are reported. The map position of the oligonucleotide (r3) used in this study for primer extensions is also reported (see text for details). ARS, autonomously replicating sequence.
Multiple-Round Primer Extension and Detection of DNAse I Footprinting
Genomic DNA (0.1-0.2 μg) was reacted with 5 U of Vent polymerase and 100,000 cpm of end-labeled oligonucleotide (specific activity, 1–2 μCi/pmol). The samples were cycled five times through the following steps: 95°C for 5 min, 69°C for 10 min, and 76°C for 3 min. The extension products were extracted with phenol, precipitated with ethanol, dissolved in formamide and dyes, and analyzed in 6% denaturing polyacrylamide gel. The DNAse I footprints were detected by autoradiography.
RESULTS
Genetic and biochemical studies in S. cerevisiae have demonstrated that a complex transcription machinery is assembled at the rDNA locus on the RNA polymerase I promoters (Nomura, 1998). Our previous studies using in vivo footprinting (Vogelauer et al., 1998) revealed that the DNA regions, shown by other approaches to be bound by the RNA polymerase I transcription factors (Nomura, 1998), are actually protected from the DNAse I digestion and that differences in binding exist when cells are grown in different conditions. To assign each footprint to each corresponding factor in vivo, we studied yeast mutants lacking specific RNA polymerase I transcription factors. We asked the following questions: Are the footprints on the CE and on the UPE maintained in CF or UAF mutants? Are the footprints sensitive to the presence of a nonfunctional RNA polymerase I and to the absence of essential transcription factors such as Rrn3p? Does a hierarchy exist in vivo in the transcription factors assembly?
Making use of the in vivo footprinting technique we are able to answer these questions and have determined an in vivo hierarchy of RNA polymerase I transcription factors.
Overview of the Promoter
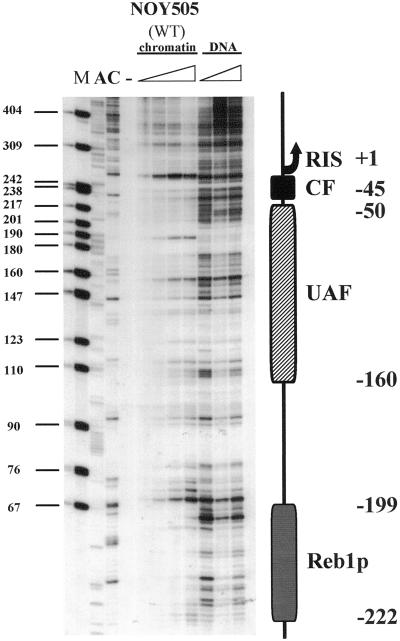
All experiments reported below were performed by digesting with DNAse I nuclei from yeast strains differing in RNA polymerase I transcription efficiency because of a lack of specific transcription factors or RNA polymerase I defects. Figure 2 shows the footprints in the wild-type (WT) strain NOY505. The whole region was analyzed by primer extension of the oligonucleotide r3 (see scheme in Figure 1 for relative position). DNA from WT nuclei treated with increasing amounts of DNAse I (NOY505) was compared with a deproteinized genomic DNA, also treated with increasing amounts of DNAse I (DNA). The observed footprints reflect the protection of the putative binding regions for the CF, UAF, and Reb1 factors. A graphical representation is also reported in Figure 2.
Figure 2.
In vivo footprinting of 35S RNA promoter. Different amounts (triangles) of DNAse I (3, 6, 12, 24 U) were introduced into purified nuclei of WT (NOY505) cells. The digestion profiles are compared with naked, deproteinized DNA (DNA) treated in vitro with different amounts of DNAse I (01, 0.2, 0.4 U). M, size marker (pBR322/MspI); lanes A and C, sequencing lanes. The schematic drawing indicates the area protected by the CF, the UAF, and Reb1p. Numbering starts from the RIS (+1).
Analyses of Mutant Strains Affected in RNA Polymerase I Transcription
To better display the results obtained with the mutant strains studied, we report the footprints by dividing them into three separate subregions: the CE, the UPE, and the Reb1p-binding site. This subdivision allows a more detailed and comprehensive analysis of the differences existing among strains in each specific region.
Effects of Transcription Factor Deficiency
The CE Region
To study the DNA protein interactions occurring on the RNA polymerase I promoter, we examined the DNAse I sensitivity of the region encompassing the RIS and the CE (+8; −28/−38 bp from the 35S RNA initiation site; Musters et al., 1989; Kulkens et al., 1991; Keener et al., 1998) in yeast strains carrying different mutations that affect the RNA polymerase I transcription. In particular, the following strains were studied: NOY558, lacking the Rrn7p subunit of the CF complex (Keys et al., 1994); NOY699, lacking the Rrn5p subunit of the UAF complex (Keys et al., 1996).
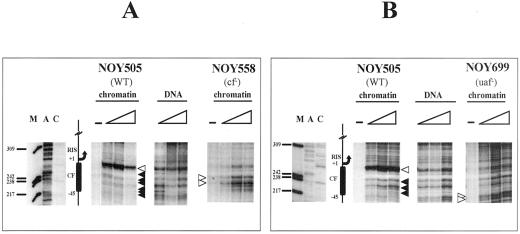
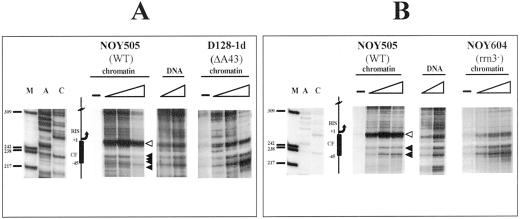
All strains mutated in these transcription components are viable in galactose medium because of the presence of an episomal copy of the 35S rRNA under the GAL7 promoter (plasmid pNOY102 or pNOY103; Nogi et al. 1991). Yeast cells were grown in YPGal to 0.4 O.D.600/ml, and nuclei were prepared (Almer and Horz, 1986) and subjected to DNAse I digestion. Figure 3 shows the comparison of the sensitivity to DNAse I digestion in the CE among three different strains: the reference WT strain (NOY505) and two mutant strains, one lacking the CF (NOY558) and one lacking the UAF (NOY699) complexes (A and B, respectively). After the in vivo enzymatic treatment with increasing amounts of DNAse I, the DNA was purified and subjected to linear amplification with Vent DNA polymerase, starting from the oligonucleotide r3 (position −267 bp, see Figure 1). The digestion profiles were compared with deproteinized DNA digested in vitro with different amounts of DNAse I and primer extended from oligonucleotide r3 (Figure 3, samples marked as DNA), which reveals the intrinsic sensitivity of naked DNA to DNAse I. The CF footprinting in the WT background is clearly visible, and it covers the area between +1 and about −45 bp, the putative binding site for the CF complex (+8; −28/−38 bp; NOY505). A major hypersensitive site (white arrowhead) marks the upstream border of the protection, and the underlying area is protected to the DNAse I digestion (black arrowheads; compare NOY505 profile with the naked DNA). Analysis of the DNAse I sensitivity of the same region in the NOY558 strain lacking the Rrn7p, an essential component of CF, allows precise attribution of the footprinting to the CF itself. In fact, the mutant sample (NOY558) treated as the WT (NOY505) does not show the same DNAse I sensitivity. The upper enhancement and the protections are lost and a different pattern of cleavage by DNAse I is observed. This result indicates a strong change in the DNA-protein interactions occurring in this region in the absence of the Rrn7p component of the CF complex. This finding indicates that a functional CF is required to produce a clear in vivo footprinting on its putative binding region.
Figure 3.
Analysis of the CE region: comparison of NOY505 (WT), NOY558 (CF mutant), and NOY699 (UAF mutant). Starting from purified nuclei, chromatin was digested in vivo with 0, 6, 12, and 24 U of DNAse I, for 5 min at 0°C and in vitro and after deproteinization (DNA) with 0.1, 0.2, and 0.4 U of DNAse I for 2 min at 0°C; after DNA purification the samples were primer extended with the labeled oligonucleotide r3. (A) Comparison of WT (NOY505) and CF mutant (NOY558). DNA, in vitro treated samples; NOY505 and NOY558, in vivo treated samples. (B) Comparison of WT (NOY505) and UAF mutant (NOY699). DNA, in vitro treated samples; NOY505 and NOY699, in vivo treated samples. M, size marker (pBR322/MspI); lanes A and C, sequencing lanes. The schematic drawing indicates the CF putative binding site and relative map positions. Protected areas are shown by black arrowheads; enhanced cleavages are shown by white arrowheads; triangles indicate increasing amounts of DNAse I in the treatment; −, untreated DNA.
To evaluate the relevance of a functional UAF complex for the binding of CF, we studied the DNAse I sensitivity of the CE in a strain lacking UAF (NOY699, lacking Rrn5p). Also in this case, we were not able to detect the CF footprinting (Figure 3B, NOY699) when compared with the WT and the naked DNA profiles. In this case, the hypersensitive site to DNAse I digestion (NOY505, white arrowhead) is missing; this suggests that the hypersensitive site is related to the presence of CF. In addition the whole CE region is clearly accessible to DNAse I cleavages. Moreover, this result shows that, even in the presence of a functional CF complex, no footprinting on CE is observed when UAF is absent.
The UPE
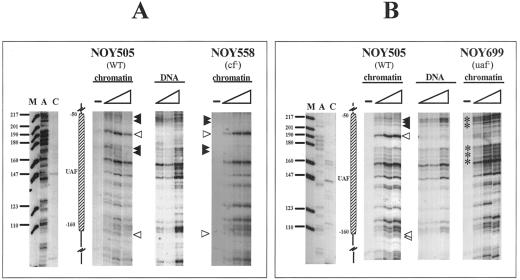
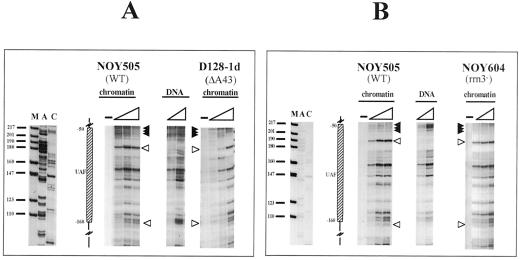
We further extended our investigation to the UPE to analyze whether any differences are detectable when crucial transcription factors for RNA polymerase I (CF or UAF) are missing. Figure 4 shows the results for the UPE region located between −41/−51 and −146/−155 bp from the RIS, where UAF supposedly binds (Musters et al., 1989; Kulkens et al., 1991; Keys et al., 1996). In Figure 4A samples treated with different amounts of DNAse I in WT (NOY505) and cf− (NOY558) nuclei are reported. Comparison with the pattern obtained by digestion of naked DNA in both strains reveals a clear footprint. In fact, enhancements of DNAse I cleavages, compared with the profile obtained with naked DNA, are visible (white arrowheads) at −70 and −160 bp, and protected areas (black arrowheads at positions −50/−70 and −90/−100 bp) are also observed. The mapping of UAF complex is, in addition, consistent with in vitro DNAse I protection experiments: using purified UAF and promoter DNA fragments, it has been found that UAF protects a region of rDNA promoter from approximately −110 to −45 bp, with a hypersensitive site at approximately −76 bp (Masayasu Nomura, personal communication). To attribute the footprint to the UAF complex, we analyzed a mutant strain lacking the Rrn5p (Figure 4B, NOY699). In this case, it is possible to detect a loss of the footprint when the DNAse I digestion profile is compared with the WT or with the CF mutant strain (see below); the hypersensitive sites at positions −70 and −160 bp, clearly evident in the WT and cf− strains (NOY505 and NOY558, respectively, Figure 4A), are completely missing in the UAF-deficient strain. Positions −50/−70 and −90/−100 bp become clearly accessible when UAF is missing (Figure 4B, asterisks). The results are consistent with the presence of an intact UAF complex, even in the absence of CF. In particular, the band mapping at −70 bp is diagnostic of the presence of UAF (see Figure 3) (white arrowheads). Also a weaker hypersensitive site is observed at −160 bp; this band marks the downstream border of the UAF complex.
Figure 4.
Analysis of the UPE region: comparison of NOY505 (WT), NOY558 (CF mutant), and NOY699(UAF mutant). Samples were treated as in Figure 3. (A) Comparison of WT (NOY505) and the CF mutant (NOY558). DNA, in vitro treated samples; NOY505 and NOY 558, in vivo treated samples. (B) Comparison of WT (NOY505) and the UAF mutant (NOY699). DNA, in vitro treated samples; NOY505 and NOY 699, in vivo treated samples. M, size marker (pBR322/MspI); lanes A and C: sequencing lanes. The schematic drawing indicates the UAF putative binding site and relative map positions. Protected areas are shown by black arrowheads; enhanced cleavages are shown by white arrowheads; triangles indicate increasing amounts of DNAse I in the treatment; −, untreated DNA.
Summarizing the data obtained from the transcription factor mutants, we can conclude that the footprinting in the CE region is observed only in the WT strain (NOY505), whereas the protections are lost in both the CF and the UAF mutants (NOY558 and NOY699, respectively). As far as the UPE is concerned, the footprinting is observed in the WT and CF mutant strains and is lost in the UAF mutant strain. These results are consistent with a hierarchy of binding for the UAF and CF complexes, suggesting that the presence of UAF facilitates binding of CF, whereas a lack of CF does not affect the UAF binding (see DISCUSSION).
Effects of a Nonfunctional RNA Polymerase I
The CE Region
To better understand the dynamics of rDNA transcription and factor binding in the previously shown regions, we studied two additional mutants in which RNA polymerase I function is suppressed, namely, the D128-1d strain, lacking the subunit A43 of the RNA polymerase I (Thuriaux et al., 1995) and the NOY604 strain lacking Rrn3p, a factor essential for RNA polymerase I transcription (Yamamoto et al., 1996). When the strain D128-1d treated in vivo with DNAse I was analyzed (Figure 5A, D128-1d), we again observed the loss of the CF footprinting, as is evident by comparison with the WT profile (Figure 5A, NOY505). Hypersensitivity and protections (white and black arrowheads, respectively) appear profoundly different. This result suggests that when a functional RNA polymerase I is missing the efficient binding of CF is also lost, the footprinting being very similar to that obtained in the absence of the CF complex (see Figure 3A, NOY558). We then asked whether in the absence of the Rrn3p a difference in the DNAse I accessibility to DNA exists. In Figure 5B, the analysis of the NOY604 strain lacking Rrn3p is reported. As is the case with D128-1d, in this strain the footprinting due to the CF complex is also lost (Figure 5B, NOY604). The diagnostic band in the upper part of the footprinting (around position +1) is clearly weakened, and the underlying area is more accessible to DNAse I in comparison with the WT profile.
Figure 5.
Analysis of the CE region: comparison of NOY505 (WT), D128-1d (RPA43 mutant), and NOY604 (RRN3 mutant). Samples were treated as in Figure 3. (A) Comparison between WT (NOY505) and RPA43 mutant (D128-1d). DNA, in vitro treated samples (0.1 and 0.2 U of DNAse I); NOY505 and D128-1d, in vivo treated samples. (B) Comparison of WT (NOY505) and RRN3 mutant (NOY604). DNA, in vitro treated samples (0.1 and 0.2 U of DNAse I); NOY505 and NOY 604, in vivo treated samples. M, size marker (pBR322/MspI); lanes A and C: sequencing lanes. The schematic drawing indicates the CF putative binding site and relative map positions. Protected areas are shown by black arrowheads; enhanced cleavages are shown by white arrowheads; triangles indicate increasing amounts of DNAse I in the treatment; −, untreated DNA.
The UPE
We next investigated whether the transcription dynamics may interfere with UAF binding to the UPE. Therefore, we analyzed the footprinting on the UAF-binding site in the same mutants, D128-1d and NOY604. When nuclei from D128-1d were treated with DNAse I (Figure 6A, D128-1d), a very similar pattern is observed with respect to the WT (Figure 6A, NOY505). The footprinted area encompassing the region from −50 to −160 bp is the same in both cases, with diagnostic hypersensitive bands (white arrowheads) positioned at −70 and −160 bp. A very similar profile is obtained with mutant cells lacking the Rrn3p (Figure 6B, NOY604). Also in this case, the protected area is characterized by two hypersensitive sites (white arrowheads) and by a region protected from DNAse I cleavage (black arrowheads). The clear similarity among NOY505, D128-1d, and NOY604 indicate that the UAF binding is not affected by the presence (WT, efficient transcription) or absence (D128-1d, NOY604, transcription deficient) of a functional RNA polymerase I. The data concerning the transcription activity and the UAF footprinting indicate that this factor remains bound to DNA also when transcription does not occur.
Figure 6.
Analysis of the UPE region: comparison of NOY505 (WT), D128-1d (RPA43 mutant), and NOY604 (RRN3 mutant). Samples were treated as in Figure 3. (A) Comparison between WT (NOY505) and RPA43 mutant (D128-1d). DNA, in vitro treated samples (0.1 and 0.2 U of DNAse I); NOY505 and D128–1d, in vivo treated samples. (B) Comparison of WT (NOY505) and RRN3 mutant (NOY604). DNA, in vitro treated samples (0.1 and 0.2 U of DNAse I); NOY505 and NOY604, in vivo treated samples. M, size marker (pBR322/MspI); lanes A and C: sequencing lanes. The schematic drawing indicates the UAF putative binding site and relative map positions. Protected areas are shown by black arrowheads; enhanced cleavages are shown by white arrowheads; triangles indicate increasing amounts of DNAse I in the treatment; −, untreated DNA.
Additional Factors: Analysis of the Reb1p-binding Site
In our previous investigations, we showed (Vogelauer at al., 1998) a footprinting on the putative binding site of the Reb1 protein at positions −222/−199 bp from the RIS. This result is confirmed also for NOY505 cells grown in galactose medium (Figure 2). Moreover we reported the binding and activity of DNA topoisomerase I at position −171 bp from the RIS (Vogelauer et al., 1998; Vogelauer and Camilloni, 1999). This latter protection is not detectable when cells are grown in a medium containing galactose as carbon source (see Figure 2).
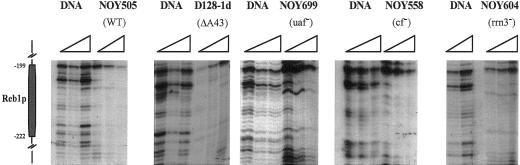
We further explored, in terms of binding to DNA, the correlation between these factors (Reb1p and DNA topoisomerase I) and the different genetic contexts that we have analyzed so far. The data reported in Figure 7 indicate that, in all five strains studied, the putative binding site for the Reb1 protein (∼220 bp upstream of the transcription start) is always protected from DNAse I digestion, when compared with naked DNA. This result suggests that the Reb1p binding to its cognate sequence is not affected by the component of the transcription machinery or by the transcriptional dynamics.
Figure 7.
Analysis of the putative Reb1p binding site. DNA, samples were treated in vitro with increasing amounts of DNAse I (as in Figure 3). NOY505, D128-1d, NOY699, NOY558, and NOY604 samples were treated in vivo (as in Figure 3) with increasing amounts of DNAse I (triangles) in different mutant strains. The schematic drawing indicates the putative Reb1p-binding region.
As far as DNA topoisomerase I is concerned, we do not observe footprinting for this enzyme in any of the strains. Actually, as we reported previously (Vogelauer et al., 1998), this footprinting is strictly dependent on the composition of the culture medium. The strains analyzed in this work were all grown in galactose, and these different growth conditions may explain the lack of footprinting previously revealed in a glucose-containing medium.
DISCUSSION
The aim of this work is to investigate the DNA-protein interactions occurring in vivo on the 35S rDNA promoter in S. cerevisiae when transcription is altered by mutations affecting either transcription factors or RNA polymerase I functioning. The in vivo footprinting methodology reveals the identity of protected regions (by using specific mutants) and recognizes mutual interactions among factors (when specific component are missing).
Identification of Specific Footprints
The interactions among RNA polymerase I transcription factors UAF, CF, and Rrn3p and the transcriptional machinery have been shown in great detail, both in genetic assays and in purified systems (Nomura, 1998; Reeder, 1999). In addition, we reported an in vivo footprinting analysis of the 35S RNA promoter showing protections lying in the putative binding regions of these factors (Vogelauer et al., 1998). The first question we asked in this work concerns the identification of those footprints to assign specific factors to each protected region. To clarify this issue we have studied mutant strains lacking specific factors by in vivo footprinting and compared the profiles with a WT strain (NOY505). When we looked at the CE in the NOY505 strain, we observed a clear footprint (compare NOY505 samples with DNA in Figure 2) in the area defined as CE. When a strain lacking Rrn5p, a subunit of the CF, is analyzed (Figure 3A, samples NOY558), the footprinting is lost and the DNAse I gains access to DNA compared with WT (Figure 3A, NOY505 samples). Because in the strain defective for a functioning CF no protections are detectable in the CE region (Figure 4A, NOY558), we conclude that the protection in this area is due to the CF complex. We also analyzed the CE when Rrn7p, a component of the UAF complex, is missing (NOY699). Also in this case (Figure 3B, NOY699 samples) a loss of protection is observed when the profile is compared with the WT (Figure 3B, NOY505 samples). We conclude that, even in the presence of a functioning CF (strain NOY699), its binding to the cognate sequence is prevented in the absence of UAF. This fact is in agreement with the reported observations (Steffan et al., 1996), according to which the recruitment of the CF on the CE in vitro is dependent on the UAF binding; therefore, in a strain lacking the UAF the CF footprinting is expected to be lost. More recently Reeder's group (Aprikian et al., 2000) showed that the overexpression of TBP in strains UAF deficient (rrn5) stimulates CF-dependent transcription at nearly the WT level in vivo; this is also shown in vitro. Furthermore, these observations suggest that binding of the CF to the CE requires the UAF or a high level of TBP.
We next analyzed the UPE to identify the UAF footprinting. In Figure 2 we show the WT UAF footprint on the UPE. However, most of the protections toward DNAse I in the promoter area (both in the CE and UPE) are less pronounced when compared with those reported in our previous investigation (Vogelauer et al., 1998). In the present study all strains used were grown in galactose medium, whereas previously we used another WT strain grown in a glucose-containing medium. We hypothesize that the difference in the intensity of the footprints is due to the growth rate (Vogelauer et al., 1998); the incomplete protection could be due to a partial occupancy of the binding factors on the repeated units. This interpretation is in agreement with the data indicating that only a fraction of the units is transcriptionally active (Dammann et al., 1993). In the WT and NOY558 (CF deficient) strains (Figure 4A), the UPE is protected and also a diagnostic hypersensitive site at −70 bp from the RIS is present; moreover, the region at −50/−70 bp is clearly protected from DNAse I digestion (black arrowheads). Conversely, the DNAse I can freely gain access to this region in the NOY699 (UAF deficient) strain (Figure 4B, samples NOY699), and the specific enhancement of cleavage is lost. This observation indicates that the footprinting on the the UPE region is not influenced by the absence of the CF (NOY558). We can therefore unambiguously attribute the identity of the in vivo protection reported on the UPE to UAF. The in vitro mapping of the UAF complex indicates protection of a region of rDNA promoter from approximately −110 to −45 bp with a hypersensitive site at approximately −76bp (Masayasu Nomura, personal communication).
Transcriptional Dynamics
The analysis of the protections from DNAse I digestion has been extended to two additional mutants in which a functioning RNA polymerase I is lacking. The first strain lacks the A43 subunit of RNA polymerase I and the second is defective in the Rrn3p, an essential component known to bind RNA polymerase I (NOY604, Yamamoto et al., 1996; Keener et al., 1998). When the strain D128-1d is analyzed (Figure 5A) we observe loss of footprinting on the CE (compare with the same region on WT, Figure 5A, NOY505 samples). Also, in the NOY604 strain lacking the Rrn3p, the protection on the CE area is lost. We conclude that the CF binding to DNA is stabilized by the presence of a functional RNA polymerase I (WT and in the presence of the Rrn3p). Most likely, the correct RNA polymerase I activity stabilizes the CF binding on the CE. The study of the protections on the UPE in the D128-1d and NOY604 strains (Figure 6) reveals that the UAF footprinting is maintained even if the RNA polymerase I is missing or lacking the Rrn3p component. This shows that binding of the UAF is independent of the presence of a functional RNA polymerase I. These data are consistent with the observations reported on binding efficiency of purified factors that demonstrated the higher affinity of the UAF for DNA compared with the CF (Keys et al., 1996; Steffan et al., 1996). Based on our results we can also suggest that the final binding of the CF to DNA is conditioned by the RNA polymerase transcription machinery.
RRN3 Function
The RRN3 gene product has been reported to be an essential component of the RNA polymerase I transcriptional apparatus (Yamamoto et al., 1996). Two hypotheses have been formulated about its function (Nomura, 1998): it may work as an RNA polymerase I recruitment factor (alike transcription initiation factor IC [TIF-IC]; Schnapp et al., 1994) or as an elongation factor. Our data are consistent with a role for Rrn3p as a recruitment factor, based on the observations that the CF footprinting in NOY604 is lost. Being the complete footprinting of the CF on the CE diagnostic of an efficient and completely assembled transcription complex, we suggest for the NOY604 strain an incomplete recruitment of factors. However, additional experimental data are needed to support this hypothesis.
Reb1 Binding
In all strains analyzed we observed a constant footprinting (Figure 7) on the Reb1p putative binding site lying ∼200 bp upstream of the 35S RNA start site. Although we cannot confirm by mutant analysis the identity of this footprinting, because strains defective in the Reb1p are not viable, we can conclude that none of the mutations affecting transcription by RNA polymerase I studied in this work has an effect on the protection observed; this indicates a nondirect involvement of this factor in RNA polymerase I transcription. This conclusion is consistent with the data reported on the transcription efficiency of strains carrying a mutation in Reb1p-binding site; the transcription rate of RNA polymerase I in these conditions is lowered by a factor of two (Kulkens et al., 1992), confirming the nonessential role of Reb1p for RNA polymerase I transcription.
We conclude that in vivo the CF and UAF bind the CE and UPE, respectively, as revealed by loss of protection to DNAse I in the corresponding mutant strains. Furthermore, our analysis shows that among factors a binding hierarchy exists, being the UAF presence essential for the CF binding but not vice versa. The data shown are in agreement with previously reported genetic and biochemical studies (Nomura, 1998). In addition, evidence is provided for a role of RNA polymerase I in affecting the CF binding. In addition, the assembly of a complete and efficient transcriptional apparatus is relevant for the CF binding, this factor being stabilized on the CE only in conditions in which transcription occurs at a high rate (Vogelauer et al., 1998). According to the last observation we consider the CF the last element required to turn the preinitiation complex into the active transcribing complex.
ACKNOWLEDGMENTS
We thank Dr. M. Nomura for providing the yeast strains and for the critical reading of the manuscript and Dr. P. Thuriaux for providing the strain D128-1d. We are also grateful to E. Di Mauro for helpful discussion and M. Caserta and S. Venditti for the critical reading of the manuscript. This work was supported by a contribution of “Istituto Pasteur Fondazione Cenci Bolognetti,” Universita' di Roma “La Sapienza”; by project ′Dinamica della cromatina nell'espressione genica' MURST, 1999; and by Consiglio Nazionale delle Ricerche Target Project on Biotechnology.
REFERENCES
- Almer A, Horz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986;5:2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprikian P, Moorefield B, Reeder RH. TATA binding protein can stimulate core-directed transcription by yeast RNA polymerase I. Mol Cell Biol. 2000;20:5269–5275. doi: 10.1128/mcb.20.14.5269-5275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R, Lucchini R, Koller T, Sogo JM. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion EA, Warner JR. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J, Josaitis CA, Dodd JA, Nomura M. Reconstitution of yeast RNA polymerase I transcription in vitro from purified components: TATA-binding protein is not required for basal transcription. J Biol Chem. 1998;273:33795–33802. doi: 10.1074/jbc.273.50.33795. [DOI] [PubMed] [Google Scholar]
- Keys DA, Lee BS, Dodd JA, Nguyen TT, Vu L, Fantino E, Burson LM, Nogi Y, Nomura M. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 1996;10:887–903. doi: 10.1101/gad.10.7.887. [DOI] [PubMed] [Google Scholar]
- Keys DA, Vu L, Steffan JS, Dodd JA, Yamamoto RT, Nogi Y, Nomura M. RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae. Genes Dev. 1994;8:2349–2362. doi: 10.1101/gad.8.19.2349. [DOI] [PubMed] [Google Scholar]
- Kief DR, Warner JR. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol. 1981;1:1007–1115. doi: 10.1128/mcb.1.11.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkens T, Riggs DL, Heck JD, Planta RJ, Nomura M. The yeast RNA polymerase I promoter: ribosomal DNA sequences involved in transcription initiation and complex formation in vitro. Nucleic Acids Res. 1991;19:5363–5370. doi: 10.1093/nar/19.19.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkens T, van der Sande CA, Dekker AF, van Heerikhuizen H, Planta RJ. A system to study transcription by yeast RNA polymerase I within the chromosomal context: functional analysis of the ribosomal RNA enhancer and the RBP1/REB1 binding sites. EMBO J. 1992;11:4665–4674. doi: 10.1002/j.1460-2075.1992.tb05568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalo D, Steffan JS, Dodd JA, Nomura M. RRN11 encodes the third subunit of the complex containing Rrn6p and Rrn7p that is essential for the initiation of rDNA transcription by yeast RNA polymerase I. J Biol Chem. 1996;271:21062–21067. doi: 10.1074/jbc.271.35.21062. [DOI] [PubMed] [Google Scholar]
- Lin CW, Moorefield B, Payne J, Aprikian P, Mitomo K, Reeder RH. A novel 66-kilodalton protein complexes with Rrn6, Rrn7, and TATA-binding protein to promote polymerase I transcription initiation in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:6436–6443. doi: 10.1128/mcb.16.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow BE, Johnson SP, Warner JR. The rRNA enhancer regulates rRNA transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1283–1289. doi: 10.1128/mcb.13.2.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W, Knol J, Maas P, Dekker AF, van Heerikhuizen H, Planta RJ. Linker scanning of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1989;17:9661–9678. doi: 10.1093/nar/17.23.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y, Vu L, Nomura M. An approach for isolation of mutants defective in 35S ribosomal RNA synthesis in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:7026–7030. doi: 10.1073/pnas.88.16.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M. Transcription factors used by Saccharomyces cerevisiae RNA Polymerase I and mechanism of initiation. In: Paule MR, editor. Transcription of Ribosomal Genes by Eukaryotic RNA Polymerase I. Austin, TX: Landes Bioscience; 1998. [Google Scholar]
- Petes TD. Yeast ribosomal DNA genes are located on chromosome XII. Proc Natl Acad Sci USA. 1979;76:410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder RH. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog Nucleic Acid Res Mol Biol. 1999;62:293–327. doi: 10.1016/s0079-6603(08)60511-5. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schnapp G, Schnapp A, Rosenbauer H, Grummt I. TIF-IC, a factor involved in both transcription initiation and elongation of RNA polymerase I. EMBO J. 1994;13:4028–4035. doi: 10.1002/j.1460-2075.1994.tb06719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MC, Choe SY, Reeder RH. In vitro definition of the yeast RNA polymerase I enhancer. Mol Cell Biol. 1993;13:2644–2654. doi: 10.1128/mcb.13.5.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Lawrence C. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1983. [Google Scholar]
- Steffan JS, Keys DA, Dodd JA, Nomura M. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 1996;10:2551–2563. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Keys DA, Vu L, Nomura M. Interaction of TATA-binding protein with upstream activation factor is required for activated transcription of ribosomal DNA by RNA polymerase I in Saccharomyces cerevisiae in vivo. Mol Cell Biol. 1998;18:3752–3761. doi: 10.1128/mcb.18.7.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuriaux P, Mariotte S, Buhler JM, Sentenac A. Gene RPA 43 in Saccharomyces cerevisiae encodes an essential subunit of RNA polymerase I. J Biol Chem. 1995;270:24252–24257. doi: 10.1074/jbc.270.41.24252. [DOI] [PubMed] [Google Scholar]
- Vogelauer M, Camilloni G. Site-specific in vivo cleavages by DNA topoisomerase I in the regulatory regions of the 35 S rRNA in Saccharomyces cerevisiae are transcription independent. J Mol Biol. 1999;293:19–28. doi: 10.1006/jmbi.1999.3154. [DOI] [PubMed] [Google Scholar]
- Vogelauer M, Cioci F, Camilloni G. DNA protein-interactions at the Saccharomyces cerevisiae 35S rRNA promoter and in its surrounding region. J Mol Biol. 1998;275:197–209. doi: 10.1006/jmbi.1997.1451. [DOI] [PubMed] [Google Scholar]
- Waldron C, Lacroute F. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J Bacteriol. 1975;122:855–865. doi: 10.1128/jb.122.3.855-865.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto RT, Nogi Y, Dodd JA, Nomura M. RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J. 1996;15:3964–3973. [PMC free article] [PubMed] [Google Scholar]