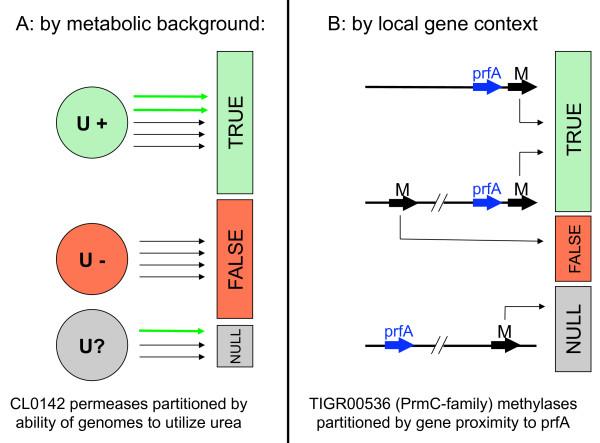
Figure 2.
Methods for partitioning a protein family for SIMBAL analysis. A) Partitioning by metabolic background assertions: members are assigned to the TRUE branch based on the observed presence of a metabolic pathway (or markers thereof) in their parent genomes (here, the ability to metabolize urea). The TRUE branch will contain both members involved in the chosen metabolism (green arrows) and, if there are many family members per genome, members which are not (black arrows). The FALSE partition will have few mis-attributed members. B) Partitioning by local genomic context: members are assigned to the TRUE branch by virtue of their proximity to a chosen marker (here, the substrate of a particular methylase among several in the larger family), while other members from the same genome are assigned to the FALSE branch. In genomes where no proximity cues are present members are omitted from the analysis. While not applicable to the PrmC example, one could also assign to the FALSE partition based on the proximity of members to markers of functions other than the one of interest.