Abstract
The Escherichia coli 3-methyladenine DNA glycosylase II protein (AlkA) recognizes a broad range of oxidized and alkylated base lesions and catalyzes the hydrolysis of the N-glycosidic bond to initiate the base excision repair pathway. Although the enzyme was one of the first DNA repair glycosylases to be discovered more than 25 years ago, and there are multiple crystal structures, the mechanism is poorly understood. Therefore, we have characterized the kinetic mechanism for the AlkA-catalyzed excision of the deaminated purine, hypoxanthine. The multiple turnover glycosylase assays are consistent with Michaelis-Menten kinetics. However, under single turnover conditions that are commonly employed to study other DNA glycosylases, we observe an unusual biphasic protein saturation curve. Initially the observed rate constant for excision increases with increasing AlkA protein, but at higher concentrations of protein the rate constant decreases. This behavior can be most easily explained by tight binding to DNA ends and by crowding of multiple AlkA protamers on the DNA. Consistent with this model, crystal structures have shown the preferential binding of AlkA to DNA ends. By varying the position of the lesion, we identified an asymmetric substrate that does not show inhibition at higher concentrations of AlkA and we performed pre-steady state and steady state kinetic analysis. Unlike other glycosylases, release of the abasic product is faster than N-glycosidic bond cleavage. Nevertheless, AlkA exhibits significant product inhibition under multiple-turnover conditions, and it binds approximately 10-fold more tightly to an abasic site than to a hypoxanthine lesion site. This tight binding could help protect abasic sites when the adaptive response to DNA alkylation is activated and very high levels of AlkA protein are present.
The integrity of the genome is challenged by spontaneous reactions that modify the nucleobases of DNA. These deleterious reactions include oxidation, deamination, and a variety of alkylation reactions involving endogenous metabolites and exogenous reactive compounds. Failure to repair these many forms of DNA damage can block DNA-templated activities such as transcription and replication and lead to permanent mutations or cell death (1, 2). The base excision repair (BER) pathway corrects the majority of damaged bases. This pathway is initiated by a DNA glycosylase that searches the genome to locate the sites of damage and then catalyzes the hydrolysis of the N-glycosidic bond to release the base lesion and create an abasic site. Downstream enzymes subsequently process this abasic site and use the information on the undamaged strand to restore the original coding sequence.
Escherichia coli contains two 3-methyladenine DNA glycosylases that are encoded by the tag and alkA genes (3). Tag (3-methyladenine DNA glycosylase I) is constitutively expressed and has high specificity for 3-methyladenine. In contrast, AlkA (3-methyladenine DNA glycosylase II) is induced as part of adaptive response to alkylation stress and recognizes a diverse group of damaged bases (3, 4). Studies exploring potential substrates for AlkA have established glycosylase activity toward more than a dozen lesions, including the alkylation adducts 3mA, 7mG, and 1,N6-ethenoadenine (εA), and the deaminated bases hypoxanthine (Hx) and xanthine (5–9). Many of the alkylated nucleotides have unstable N-glycosidic bonds and they are difficult to work with, but previous work suggests that AlkA has similar rate enhancements for a wide range of disparate lesions (10). We have focused on the excision of Hx, because the homogenous substrate can be prepared and it has a stable N-glycosidic bond.
A wealth of structural information is available for the interaction of AlkA with DNA. AlkA has been crystallized in complex with a 1-azaribose transition state analog (11) and there are several structures of AlkA bound to the ends of oligonucleotides (12, 13). Most recently, engineered protein-DNA crosslinks were used to obtain structures of AlkA interacting with undamaged DNA (14). Remarkably, all of these structures show a very similar overall set of protein-DNA interactions with only subtle rearrangements that are localized to the active site pocket and neighboring residues (14).
Mechanistic insights into the AlkA reaction have lagged behind the structural studies. In the current work we have characterized the single- and multiple-turnover kinetics for excision of Hx from oligonucleotides in which the lesion is located different distances from the ends. Comparison of these substrates provides evidence that AlkA binds to DNA ends more tightly than to Hx lesions, and this restricts the binding of subsequent AlkA protamers. We identified a well-behaved substrate that is amenable to kinetic studies and describe the minimal kinetic mechanism for recognition and excision of Hx.
MATERIALS AND METHODS
Preparation of Proteins
Full-length AlkA and truncated human alkyladenine DNA glycosylase (AAG) lacking the first 79 amino acids (Δ80 AAG) were expressed in Escherichia coli and purified as described previously (10, 15). Initial estimates of the protein concentration were made based upon the UV absorbance and the calculated extinction coefficients. The concentration of active Δ80 AAG was determined from the burst amplitude for the excision of Hx, as previously described (16). The concentration of active AlkA was determined by titration with a DNA inhibitor (see below).
Preparation of Oligonucleotides
DNA substrates were obtained from Integrated DNA Technologies or the Keck Center at Yale University and purified by denaturing polyacrylamide gel electrophoresis. The DNA was then extracted and desalted by reverse phase (C18 Sep-pak, Waters). The concentrations of single-stranded DNA were determined by UV absorbance at 260 nm using the calculated extinction coefficients. For the εA-containing oligonucleotide, the extinction coefficient was calculated based on an identical sequence containing A instead of εA and a value of 9400 M−1 cm−1 was subtracted to account for the weaker absorbance of εA relative to A. Prior to glycosylase assays, the oligonucleotides were annealed with a 2-fold excess of the complementary strand by being heated to 90 °C and cooled to 4 °C over a period of 15 minutes. Control experiments have shown that the excess single strand does not affect the observed rate constants (data not shown). The sequences of the oligonucleotide duplexes are shown in Table 1.
Table 1.
Sequences of Oligonucleotide Substrates
| 25 | 5′ -(FAM) CGATAGCATCCTXCCTTCTCTCCAT-3′ |
| 3′ -GCTATCGTAGGATGGAAGAGAGGTA-5′ | |
| 19u | 5′ -(FAM) CATCCTXCCTTCTCTCCAT-3′ |
| 3′ -GTAGGATGGAAGAGAGGTA-5′ | |
| 19d | 5′ -(FAM) CGATAGCATCCTXCCTTCT-3′ |
| 3′ -GCTATCGTAGGATGGAAGA-5′ |
Oligonucleotides were annealed to give the indicated duplex substrates containing a single lesion site (X). The DNA tested includes undamaged adenosine (A), the lesions inosine (I) and 1,N6-ethenoadenosine (εA), the abasic product, and the transition state analog pyrrolidine (Y).
The abasic DNA product was formed by the complete hydrolysis of I•T DNA with AAG, followed by phenol/chloroform extraction, and buffer exchange using Sephadex G-25 that had been equilibrated with annealing buffer (10 mM NaMES, pH 6.5, 50 mM NaCl). The fraction abasic was determined to be >90%, with a small amount of nicked DNA and a small amount of intact substrate (17). The concentration of DNA after buffer exchange was determined from the fluorescence of the fluorescein label after purification.
General Glycosylase Activity Assay
Reactions were performed at 37°C with the enzyme and DNA substrates mixed in a solution containing 50 mM NaMES (pH 6.1), 1 mM EDTA, 1 mM DTT, 10% (v/v) glycerol, 0.1 mg/mL BSA. The ionic strength was adjusted to 100 mM with NaCl. The pH value of 6.1 is the optimal pH for AlkA-catalyzed excision of neutral lesions (10). Reactions were initiated with the addition of a small volume of enzyme to result in a total reaction volume of 20 µL. Aliquots were withdrawn at the desired times and quenched with equal volume of 0.4 M sodium hydroxide to attain a final concentration of 0.2 M. For long time courses, the quenched samples were stored at 4°C until all aliquots were taken to prevent the base-catalyzed ring opening and subsequent depurination of εA lesions. The abasic sites were quantitatively cleaved by heating samples at 70°C for 15 minutes. For the εA-containing substrate, control experiments showed negligible cleavage of εA sites by this heating process (≤1%, data not shown). Samples were then mixed with 2 volumes of loading buffer (98% formamide, 10 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol FF) and resolved by 20% (w/v) polyacrylamide sequencing gels containing 6.6 M urea. Gels were scanned using a Typhoon Trio fluorescence imager (GE Healthcare) with 532 nm excitation and a 526 short-pass filter to detect fluorescein. The fluorescence signal was quantified using Image Quant TL (GE Healthcare) and the fraction product for each time point was calculated (fraction product = intensity of product band/(intensity of product band + intensity of substrate band)).
To evaluate the stability of AlkA under these standard reaction conditions, AlkA was incubated in the absence of DNA for periods of time up to 50 hours, and then the steady state glycosylase activity was measured. The half-life for AlkA inactivation under these conditions was ≥100 hours (see Supporting Information).
Determination of Active AlkA Concentration
We titrated AlkA with a 25mer pyrrolidine-containing oligonucleotide duplex (0 to 400 nM) to determine the concentration of active AlkA. The enzyme was preincubated at 37°C with the inhibitor for 10 minutes before the reactions were initiated with the addition of 19mer Hx-containing substrate (19u). Reactions were performed under our standard conditions with 1 µM DNA substrate and at two different concentrations of AlkA (100 and 200 nM). The relative AlkA activity in the presence of inhibitor was determined by the ratio of the observed velocity in the presence of inhibitor divided by the velocity in the absence of inhibitor (Vobs/Vmax). The fraction of active AlkA was determined from this titration plot, using the quadratic binding equation (eq 1) in which E is the concentration of AlkA and I is the concentration of pyrrolidine-containing DNA inhibitor.
| (1) |
Single Turnover Glycosylase Activity
DNA substrates were incubated with excess enzyme to ensure single-turnover conditions. Unless otherwise indicated, the concentration of substrate was 20 nM and the concentration of AlkA was varied from 40 to 3000 nM. Samples were removed and quenched at appropriate intervals and the extent of glycosylase reaction was analyzed as described above. The fraction of base excised was fit by a single exponential according to eq 2, in which F is the fraction product, A is the amplitude, kobs is the observed single turnover rate constant, t is the reaction time, and c is the amount of pre-existing abasic DNA.
| (2) |
The hyperbolic dependence of the single turnover rate constant was fit by eq 3, in which kmax is the maximal single turnover rate constant when AlkA is fully saturated and the K1/2 is the concentration at which the enzyme is 50% saturated. The K1/2 value is analogous to a KM determined in steady state experiments and is equal to the dissociation constant (Kd) if substrate binding is readily reversible. Given the low values of kmax determined here, this is likely to be the case.
| (3) |
The single-turnover excision of Hx from 19d by AlkA was too slow to cover the entire time course without concern for the loss of stability of AlkA. Therefore, time points were restricted to the first 21 hours, which corresponds to between 40–65% completion. The experimental data were fit by eq 2 with c fixed to 0.03 (based on quantification of product band from control reactions with no enzyme) and A was fixed to a value of 0.95 (based on fraction product at completion from reaction with the same substrate and saturating concentration of AAG).
For substrates in which inhibition was observed at high enzyme concentrations, we used a simple noncompetitive inhibition model using eq 4, in which Kd is the binding constant for the stimulatory site and Ki is the binding constant for the inhibitory site on the DNA substrate. This is analogous to previous treatment for substrate inhibition in which a second substrate molecule can bind to an enzyme (18).
| (4) |
Steady State Kinetics
Steady state studies were performed with DNA substrate in 100-fold excess over enzyme. To investigate the possibility that AlkA exhibits burst kinetics, the ratio of substrate to enzyme was lowered (50:1, 25:1, and 10:1) to observe both pre-steady state and multiple-turnover kinetics in a reasonable time frame. Because significant product inhibition was observed under steady state conditions, we only used the first 8% of reaction to calculate the initial velocities (Vobs). Plots of Vobs/[E] versus substrate concentration were fit by the Michaelis-Menten equation (eq 5).
| (5) |
Measurement of Inhibition Using Steady State Kinetics
To determine the relative affinity of AlkA for undamaged DNA and for the abasic DNA product, we measured the competitive inhibition by these species under multiple turnover conditions. The advantage of this approach over direct binding assays is that all DNA species are in excess over enzyme and therefore it is unlikely that multiple enzymes can bind to the same DNA. These experiments used 2 nM AlkA, 100 nM I•T 19u substrate, and the amount of either undamaged (A•T) or abasic 25mer DNA product was varied between 10 and 1000 nM. Initial rates were measured as described above and the resulting inhibition was fit by the equation for competitive inhibition (eq 6). Under these conditions the inhibition constant (KI) is equal to the dissociation constant (Kd) for the inhibitor.
| (6) |
RESULTS
Determination of the Concentration of Active AlkA
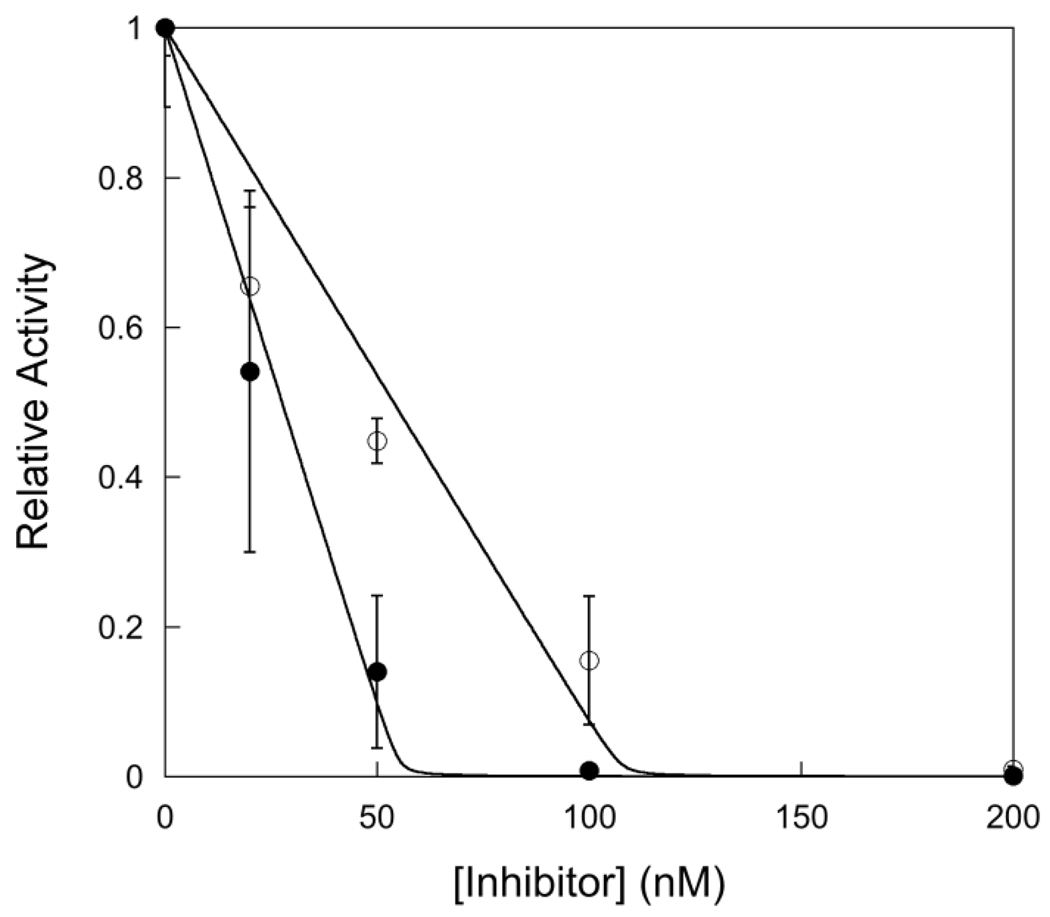
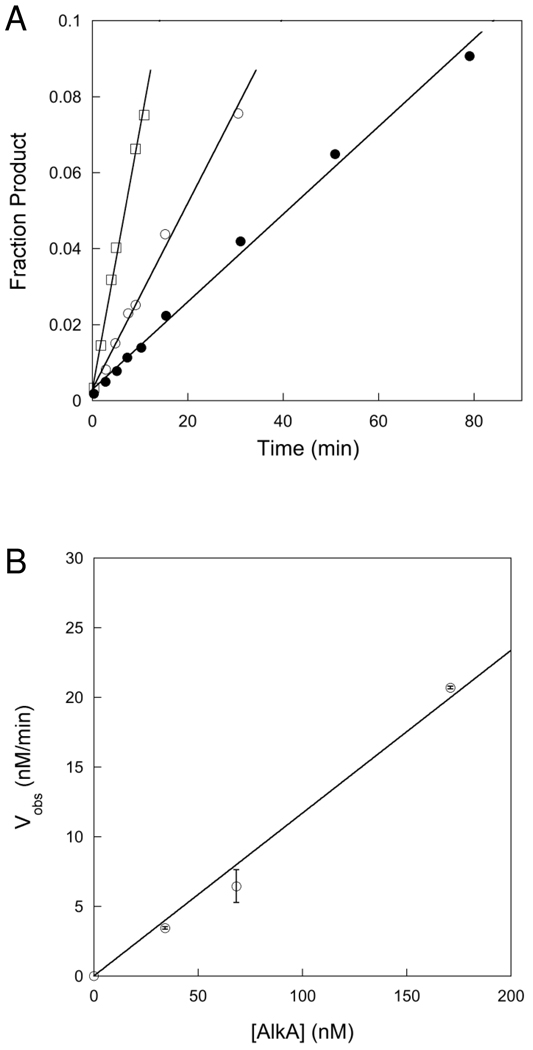
In order to compare single turnover and multiple turnover kinetics it is critical to know the concentration of active enzyme. The concentration of active sites can be determined by measuring the amplitude of a pre-steady state burst or by titrating the enzyme with a tight binding inhibitor. For the AlkA-catalyzed excision of εA and Hx no burst was observed (see below), therefore we took advantage of a previously identified tight binding inhibitor of AlkA, a pyrrolidine-containing oligonucleotide. The pyrrolidine bears a positive charge at pH values below 7 and therefore mimics the oxocarbenium ion transition state of a glycosylase reaction (19, 20). A binding affinity of ~20 pM has previously been reported for a 25mer-oligonucleotide containing a pyrrolidine site (20). AlkA was incubated with various concentrations of pyrrolidine-DNA inhibitor under conditions far above the dissociation constant for inhibitor binding, and the steady-state rate of N-glycosidic bond cleavage was determined by measuring the initial rates. As expected for a tight binding inhibitor, the concentration dependence of the inhibition followed a quadratic function that is linear at low concentrations of inhibitor (Figure 1). Titration of 2-fold higher concentration of AlkA increased the x-intercept by 2-fold, confirming that stoichiometric binding of inhibitor occurred under these conditions. These data indicate that the actual concentration of active AlkA is ~57% of the value estimated from the absorbance at 280 nm. For the remainder of the experiments that are presented we use the concentration of active AlkA that was determined from this titration.
Figure 1.
Titration with a tight-binding pyrrolidine inhibitor to determine the concentration of active AlkA. Experiments were performed using 1 µM 19mer I•T substrate (19u) with varying concentrations (from 0 to 400 nM) of 25mer pyrrolidine inhibitor (Y•T). The concentration of AlkA was 100 nM (●) and 200 nM (○). The fraction of active AlkA was determined by measuring the initial rate of product formation and plotting the relative rate (Vobs/Vmax) versus the concentration of inhibitor. The average and standard deviation of two to four replicates are shown. This titration gives an average value of 0.57 ± 0.03 for the fraction of active AlkA, assuming a single monomer binds to each DNA (11).
Single Turnover Excision of Hx
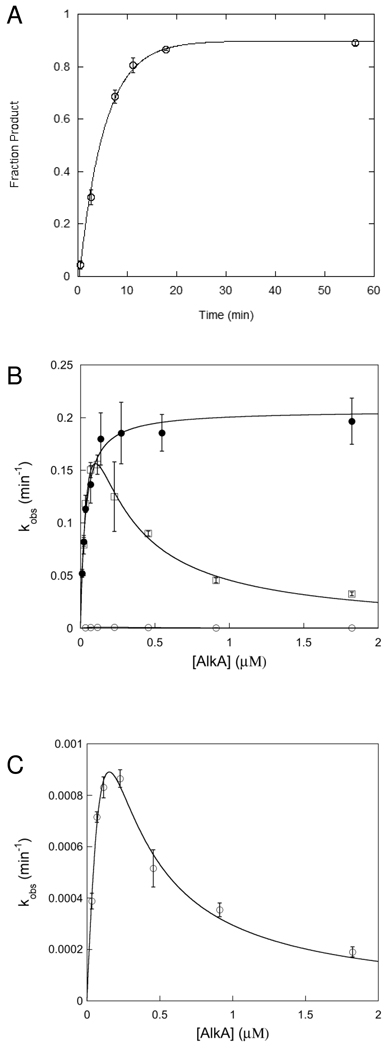
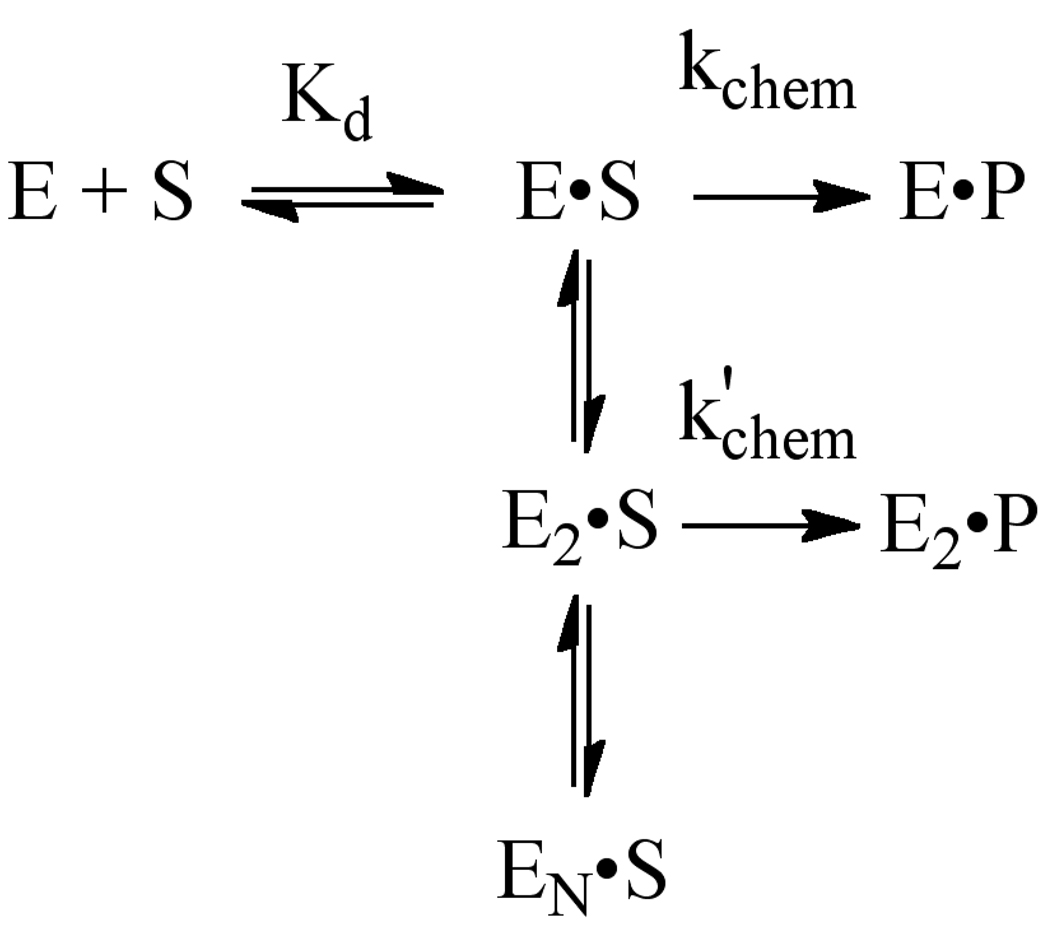
The crystallographic footprint of AlkA binding to DNA is ~8 base pairs (11). Therefore, we used a 25mer symmetric oligonucleotide substrate that was expected to be sufficiently large to support AlkA binding (Table 1). Single turnover reactions were performed with AlkA in excess over the 25mer substrate. In all cases, the reactions followed first order kinetics and the reaction progress curves were fit with single exponentials. The single turnover reaction monitors all of the steps up to and including the first irreversible step, N-glycosidic bond cleavage, and is not affected by product release or product inhibition steps. It is expected that varying the concentration of enzyme under these conditions would result in a hyperbolic dependence upon the concentration of enzyme according to eq 3. However, for the 25mer I•T substrate we observed a reproducible reduction in rate at high concentrations of AlkA (Figure 2B; squares). These data are consistent with AlkA binding to another site on the DNA that inhibits the binding or activity of AlkA at the lesion site (Scheme 1). According to this model, binding of two or more AlkA monomers could severely reduce or even completely block the activity of one monomer. As several crystal structures have been reported in which AlkA is found bound to the ends of short oligonucleotides, instead of at a central damaged site, we considered the possibility that the inhibition at high concentration of AlkA could be attributed to end binding by AlkA. Similar inhibition was observed for the AlkA-catalyzed excision of the alkyl adduct, 1,N6-ethenoadenine (εA), suggesting that the inhibition was not exclusive to the I•T form of damage (see Supporting Information). These observations suggest that AlkA has roughly comparable affinity for DNA ends as it has for internal sites of DNA damage.
Figure 2.
Single-turnover excision of Hx by AlkA. (A) Representative time course for AlkA-catalyzed excision from I•T 19u substrate (20 nM) at saturating concentration of AlkA (1.8 µM). Reactions were performed in triplicate and the average value is shown (error bars indicate the standard deviation). The reaction progress curve was fit by the equation for a single exponential (see Materials and Methods for details). (B) Dependence of the single turnover rate constant on the concentration of AlkA for I•T DNA. The symmetric 25mer substrate (□), the upstream shortened 19u (●) and the downstream shortened 19d (○) substrates are shown. Each data point is the average of at least 3 independent determinations and the error bars indicate the standard deviation. (C) The data for the 19d substrate are replotted from B to show that this substrate also shows inhibition at high concentration of AlkA. The equations for a simple hyperbolic dependence (eq 3) and for the inhibitory site model (eq 4) are given in the Materials and Methods and the values obtained from these fits are summarized in Table 2.
Scheme 1.
Model to explain inhibition by multiple molecules of AlkA
We next tested the simple idea that changing the position of the lesion relative to the end might alter the inhibition. We prepared oligonucleotides that lacked 6 base pairs from either the upstream (5’ from the lesion) or downstream (3’ from the lesion) portion of the duplex (Table 1), and measured the AlkA concentration dependence for the single turnover reaction. The results are shown in Figure 2B. Removing 6 base pairs from upstream (19u) did not result in a significant decrease in AlkA glycosylase activity, but rather eliminated the inhibition that was observed at high concentrations of AlkA for the 25mer substrate. In contrast, removal of 6 base pairs downstream from the lesion (19d) resulted in a profound decrease in AlkA activity. The decreased activity in this orientation is consistent with the asymmetric footprint of AlkA, whereby more extensive protein-DNA contacts are made downstream from the lesion site (11). Fraying of the ends is likely to exacerbate this effect (21). Nevertheless, the AlkA-catalyzed glycosylase activity on the 19d substrate showed the same inhibition at high concentrations of AlkA as was observed for the 25mer (Figure 2C).
To gain additional insight into the binding affinities of AlkA for the lesions and DNA ends, we fit the inhibition to a model in which the DNA contains multiple binding sites, with at least one inhibitory site in addition to the lesion site (eq 4, see “Materials and Methods”). This provided estimates for the maximal single turnover rate constant (kmax) and the dissociation constants for the lesion site and for the inhibitory site and these are summarized in Table 2. For the 25mer substrate, AlkA has modestly higher affinity for the lesion site than for the DNA ends. For the 19d substrate, AlkA showed much tighter affinity for the ends than for this suboptimal lesion site.
Table 2.
Kinetic Parameters for AlkA-Catalyzed Excision of Hypoxanthine
| Substrate | kmax (min−1)a | Kd (nM) | Kib (nM) | kcat (min−1) | KM (nM) | kcat/KM (M−1min−1) |
|---|---|---|---|---|---|---|
| 19u | 0.21 ± 0.01 | 32 ± 3 | – | 0.092 ± 0.003 | 52 ± 10 | 1.7 × 106 |
| 19d | 0.007 ± 0.001c | 500 ± 50c | 47 ± 5c | 0.00054 ± 0.00004 | NDd | NDd |
| 25 | 0.28 ± 0.01e | (42)e | 220 ± 30e | 0.075 ± 0.003 | 42 ± 9 | 1.8 × 106 |
The sequences of these I•T substrates are given in Table 1. All reactions were carried out at 37 °C and the reaction buffer contained 50 mM NaMES, pH 6.1, 10% (v/v) glycerol, 1 mM EDTA, 1 mM DTT, 0.1 mg/mL BSA and sufficient NaCl to obtain an ionic strength of 100 mM.
The maximal single-turnover rate constants were determined with a saturating concentration of AlkA.
Kd for the inhibitory site. The values were determined using eq 4 (Figure 2). No inhibition was observed for the 19u substrate.
At suboptimal concentrations of AlkA the single-turnover excision was too slow to follow all of the reactions to completion (See Materials and Methods for details). The errors were estimated by setting each variable to the optimal value and monitoring the uncertainty in the values that were not fixed.
ND, not determined.
As the inhibitory model contains three variables, we fixed the Kd value for this substrate to be the KM value determined under multiple turnover conditions. This constrains the inhibitory Ki value and the kmax value as reported. Letting all parameters float gave a kmax value of 0.4 min−1, a Kd value of 140 nM and a Ki value of 230 nM.
Multiple Turnover Glycosylase Activity of AlkA
To obtain a comprehensive view of the entire AlkA-catalyzed reaction, we also performed multiple turnover glycosylase assays. Under conditions with excess DNA over protein it is unlikely that multiple AlkA molecules would be present on a single DNA. However if AlkA binds to DNA ends then the steady state reaction rate is expected to be slower than the theoretical rate on a DNA without ends, because end binding is competitive with lesion binding. We found that even at saturating concentration of DNA substrate, the rate of AlkA-catalyzed multiple turnover reactions decreased significantly above 10% reaction (see Supporting Information). In principle, this lack of linearity could be due to instability of AlkA or product inhibition. Control experiments demonstrated that AlkA is stable under the conditions used (see Supporting Information), suggesting that AlkA is extremely sensitive to product inhibition. Below we test this hypothesis directly and show that the abasic DNA product is a potent inhibitor of AlkA.
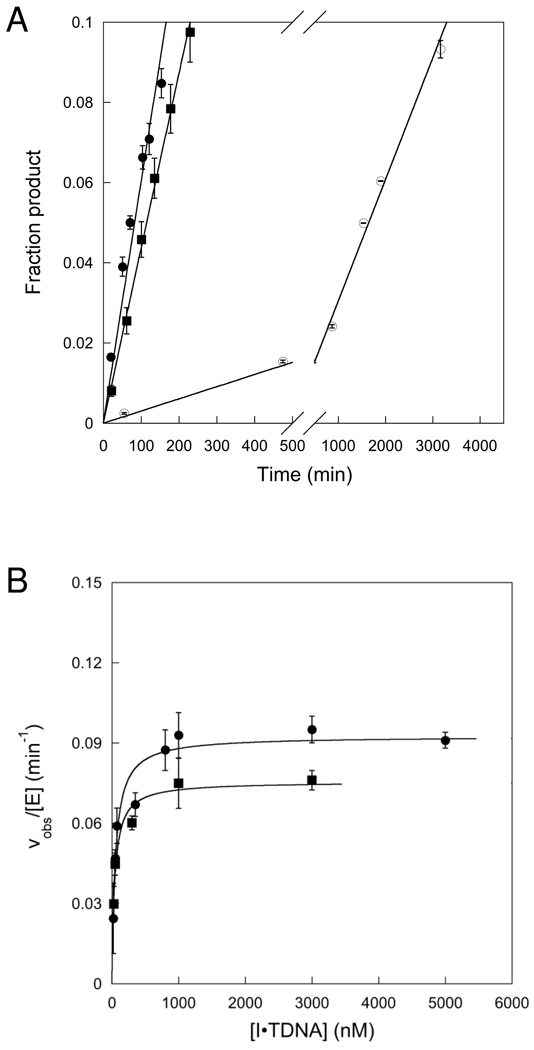
To avoid the problem of product inhibition, we monitored initial rates under conditions in which less than 8% product was formed. At less than 8% product, the reaction progress curves were linear (Figure 3A). Confident that initial rates were measured, we varied the concentration of both 25 and 19u substrates. The substrate concentration dependence is very similar for both substrates and obeys Michaelis-Menten kinetics (Figure 3B). The KM value of 52 nM that was determined for the 19u substrate is very similar to the K1/2 value of 32 nM that was determined for the single turnover reaction on this same substrate (Table 2). However, the kcat value of 0.09 min−1 is approximately half the value of kmax that was determined for the single turnover reaction. This raises the possibility that another step after N-glycosidic bond cleavage is rate-limiting for multiple turnover excision. Most previously characterized glyocosylases are limited by release of the abasic DNA product, therefore this seemed like a possible candidate (17, 22–24). We performed kinetic experiments in which the concentration of AlkA was 2–10% of the concentration of DNA. If product release was rate-limiting, then a pre-steady state burst would be expected between 2 and 10%. However, no burst was detected under these conditions (Figure 4A). Furthermore, the initial rates measured for this first turnover by AlkA were almost identical to those determined for the first 8 turnovers (steady state conditions), corresponding to a kcat value of ~0.1 min−1 for the 19u substrate (Figure 4B). Thus, the absence of a burst confirms that product release is much faster than N-glycosidic bond cleavage.
Figure 3.
Multiple-turnover glycosylase activity of AlkA. (A) Representative data for AlkA-catalyzed excision of Hx from 19u (●), 19d (○), and the symmetric 25mer (■) with 10 nM AlkA and 1000 nM DNA. (B) The concentration dependence is shown for 25 (■) and 19u (●) I•T substrates. Each data point is the average of at least 3 independent determinations and the error bars indicate one standard deviation. The lines indicate the best fit to the Michaelis-Menten equation (eq 5). The kcat and KM values are given in Table 2.
Figure 4.
AlkA-catalyzed excision of Hx under conditions suitable for detecting a burst. (A) Reactions were performed with saturating concentration of I•T 19u DNA (3 µM) and 34 (●), 68 (○), and 140 nM (□) AlkA. Reaction rates were linear, indicating the absence of a burst. (B) The initial rates from panel A are plotted as a function of the concentration of AlkA. The line shows a linear fit and yields a kcat value of 0.12 ± 0.01 min−1 that is in reasonable agreement with the kcat value of 0.092 min−1 that was determined from a wider range of substrate concentrations under steady-state conditions (Table 2).
The calculated value of kcat depends upon the concentration of active AlkA and the concentration of DNA substrate, therefore it is difficult to completely rule out the possibility that the 2-fold difference between single turnover and multiple turnover excision is due to experimental error. However, the decreased reaction rate under multiple turnover conditions is consistent with the model that AlkA binds to DNA ends with similar affinity as to base lesions. A 2-fold slower kcat value could result from an equal probability that AlkA would be nonproductively bound to the ends or productively bound to the lesion.
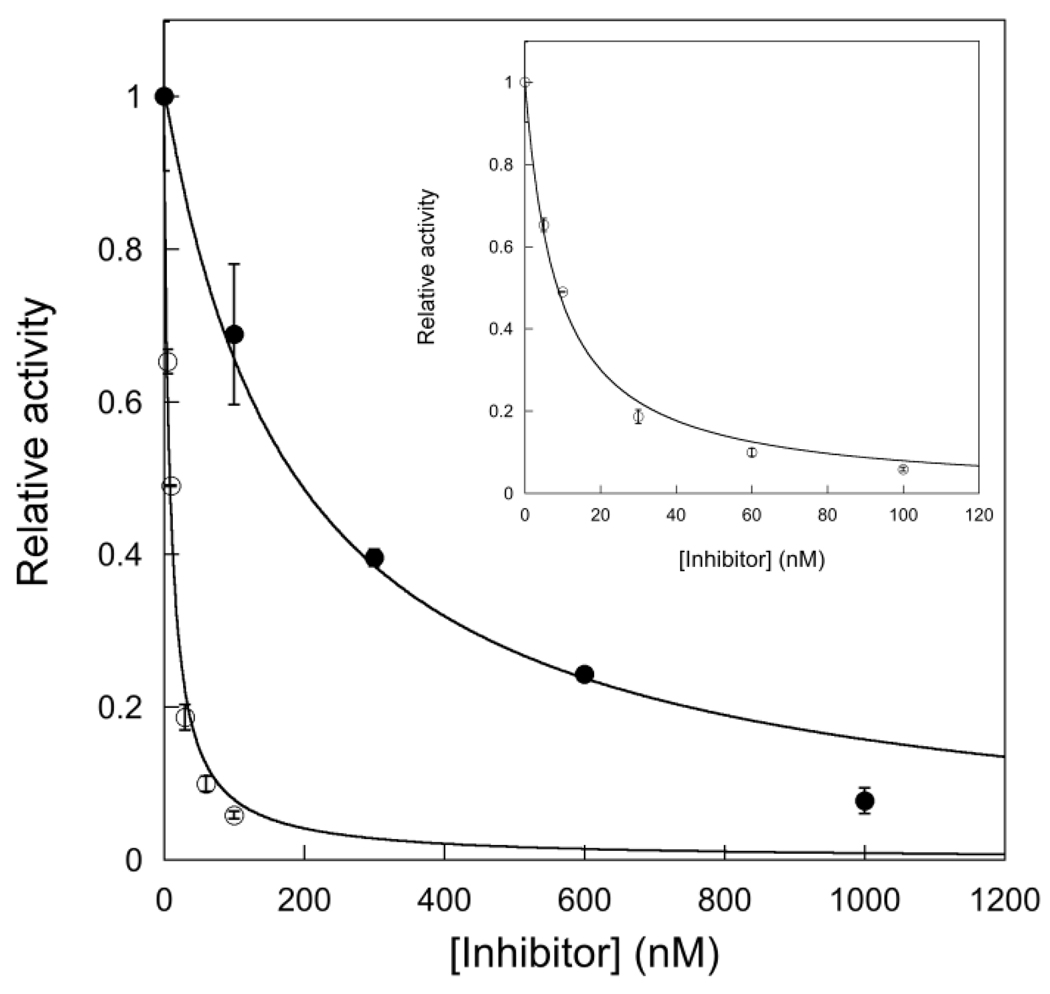
Measurement of the Dissociation Constant for Undamaged and Abasic-containing DNA
We next used steady-state kinetics to measure the inhibition of AlkA by a 25mer undamaged DNA (A•T 25) and by the abasic product. Under these conditions the inhibition constant is simply the dissociation constant (Kd) for AlkA binding. The inhibition by added DNA followed the expected equation for competitive inhibition, yielding Kd values of 61 nM for the undamaged DNA and 3 nM for the abasic DNA product (Figure 5). It is striking that the undamaged DNA binds to AlkA with very similar affinity as the I•T substrates (30–50 nM; Table 2). This suggests that features of a DNA end closely mimic features important for binding to damaged DNA, and is consistent with the very similar structures observed for binding of AlkA to DNA ends and to an internal 1-azaribose site (11–13).
Figure 5.
Inhibition of AlkA by undamaged and abasic-containing DNA. Reactions contained 100 nM I•T 19u DNA, 2 nM AlkA, and the indicated concentration of abasic or undamaged (A•T) 25mer DNA inhibitor. Lines indicate the best fits to the equation for competitive inhibition and give Kd values of 61 ± 9 nM for undamaged DNA (●) and 2.8 ± 0.3 nM for abasic product (○; see inset).
DISCUSSION
AlkA has been extensively studied and there has been recent progress in elucidating the structure of the protein in complex with DNA (11–14). Previous work has focused on identifying the mechanism of transcriptional regulation (25) and in identifying new substrates of the enzyme. These studies identify AlkA as having one of the most broad substrate ranges of any DNA repair glycosylase (5–7, 9, 10). Despite the identification of dozens of deaminated and alkylated substrates, the kinetic mechanism has not previously been reported for any substrate. Here we have used single turnover and multiple turnover kinetics and DNA binding assays to determine the minimal kinetic mechanism for the recognition and excision of Hx from I•T lesion sites. These studies provide kinetic evidence that multiple AlkA molecules can bind close together and block repair of damaged DNA.
Multiple AlkA Molecules Bind to DNA and Interfere with Repair
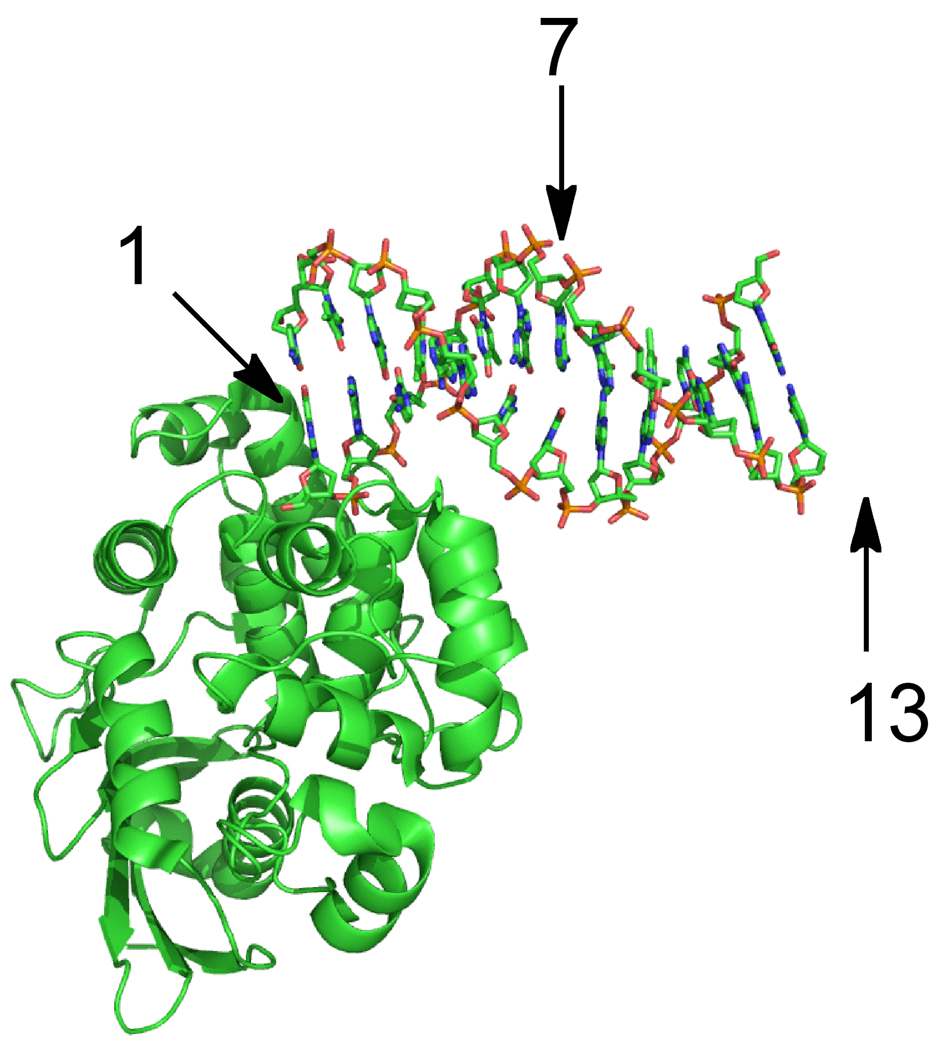
Single-turnover kinetic analysis of AlkA revealed an unusual dependence upon the concentration of protein, in which it reached an optimum concentration and then declined beyond that (25mer I•T; Figure 2B). This type of behavior could be explained by protein aggregation or by binding of multiple molecules to the oligonucleotide substrate. Numerous lines of evidence argue against a general aggregation effect, including the ability to purify and crystallize AlkA at much higher concentration of protein, and the finding that even concentrated stocks of protein are stable for months at 4 °C (data not shown). However, the most convincing evidence is that the effect is completely absent for a related substrate that bears the identical local sequence context, but is shortened asymmetrically so that the lesion is located 7 base pairs from the end, instead of 13 base pairs from the end (Figure 2B). The inhibition was retained when the opposite (downstream) end of the DNA was shortened (Figure 2C). These observations, and the finding that AlkA prefers to crystallize in an end-binding conformation (Figure 6; (12, 13)), suggest that binding by AlkA to the end of the DNA affects the binding register of additional protamers (Figure 7). We are forced to invoke multiple binding of AlkA molecules to the interior of the duplex, because the physical distance between the central position of a 25mer substrate and an AlkA molecule bound at the end of the DNA is too great to allow a direct interaction (Figure 6).
Figure 6.
Structure of AlkA bound to the end of a duplex. The figure was rendered with Pymol and the coordinates are from the Protein database (3CWT; (12)). This crystal form contains two oligonucleotides and four AlkA monomers per asymmetric unit, but only one AlkA-DNA interaction is shown. AlkA interacts predominantly with the strand that donates the 5’ end of the DNA. Arrows indicate the nucleotide position relative to the 5’end. Position 7 corresponds to the position of the lesion in the 19u substrate, and this lies on the opposite face of the DNA from a protein bound to the 5’ end. Position 13 (not present in the crystallized 12mer oligonucleotide) is on the same face as the end-bound AlkA molecule, but is separated by approximately one turn of the helix. This suggests that more than one AlkA molecule would need to bind to the 25mer symmetric substrate before the lesion site (position 13) would experience interference.
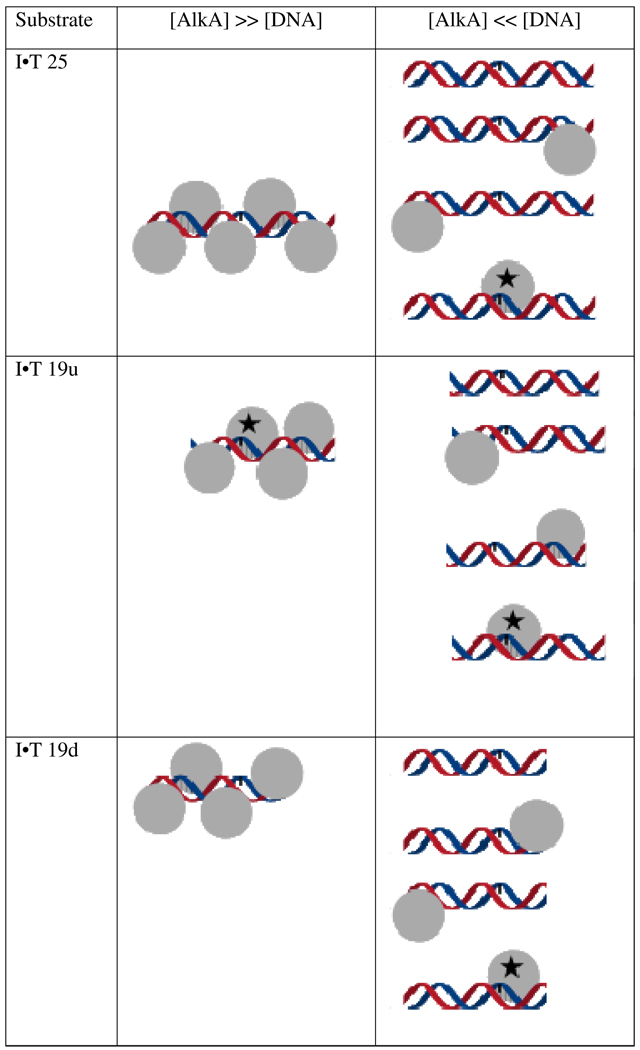
Figure 7.
Models for inhibition of repair by binding of multiple AlkA molecules. If more than one AlkA (grey spheres) is bound ([AlkA] >> [DNA]), then not all sites can be sampled by the active site and the position relative to the end determines whether or not a damaged base (rectangle) is engaged productively ( ). If there is limiting amount of AlkA ([AlkA]<<[DNA]), then there will be an ensemble of DNA with AlkA bound at different binding sites across the DNA and every site can be sampled. For simplicity, only a few possible bound states are shown.
). If there is limiting amount of AlkA ([AlkA]<<[DNA]), then there will be an ensemble of DNA with AlkA bound at different binding sites across the DNA and every site can be sampled. For simplicity, only a few possible bound states are shown.
Consistent with this hypothesis, AlkA obeys normal Michaelis-Menten kinetics under conditions of excess substrate (Figure 3). The pattern of inhibition under single turnover conditions requires that AlkA bind with greater than or equal affinity to the DNA end and to internal adjacent sites, as it binds to an I•T lesion. This tight binding was confirmed by measuring the binding of DNA that contains ends, but no sites of damage. Under these conditions the KM value for the I•T reaction of 30–50 nM (Table 2) is similar to the Kd for binding to undamaged DNA of 61 nM (Figure 5).
AlkA is induced in response to DNA damage, and expressed at very high levels. In light of these findings, it is worth considering whether the ability of AlkA to bind to broken DNA ends, and to undamaged DNA at very high density, could have physiological consequences beyond its DNA glycosylase activity. Along these lines, it is notable that several other DNA repair proteins have been shown to bind to DNA ends. For example, the single-strand selective uracil DNA glycosylase (SMUG1) and the mismatch-specific uracil DNA glycosylase (MUG) have both been crystallized in complex with DNA ends (26, 27). For MUG it has recently been noted that multiple enzyme molecules are required for binding to DNA and for the excision of uracil from synthetic oligonucleotides (28). Other repair proteins have been reported to bind in close proximity on DNA. For example, thymine DNA glycosylase (TDG) has been found to form a dimer with one molecule bound to the site of damage and the other bound in close proximity (29, 30). Biochemical characterization does not reveal any functional effect on the glycosylase reaction in this case (30). It is also intriguing that O6-alkylguanine-DNA alkyltransferase has been shown to bind at very high density to undamaged DNA (31). In all of these cases it is not clear whether these properties are a byproduct of strong nonspecific DNA binding that would facilitate a genome-wide search for DNA damage, or play some role in DNA damage signaling or in shielding a site of DNA repair.
Kinetic Mechanism for AlkA-Catalyzed Excision of Hypoxanthine
We took advantage of the fact that the glycosylase reaction catalyzed by AlkA on the 19u I•T substrate follows a simple concentration dependence to describe the minimal kinetic mechanism of AlkA. We infer that this asymmetric substrate is sufficiently long to support AlkA binding, because very similar steady state kinetic parameters were observed for this substrate and for a symmetrical 25mer substrate (Table 2). We assume that DNA binding and nucleotide flipping are rapid, because this has been the case for every glycosylase that has been previously studied (32). Regardless of whether this assumption is valid, the single turnover glycosylase reaction places a lower limit on the rate of N-glycosidic bond cleavage of 0.2 min−1. In contrast, the steady state rate constant (kcat) is approximately 2-fold lower. This could indicate a slow step after N-glycosidic bond cleavage such as product release. This was an attractive hypothesis, because slow product release has been observed for many DNA glycosylases (33). However, this model predicts the existence of a pre-steady state burst, which was not observed. This suggests that the rate constant for product release is greater than 0.2 min−1 (Figure 4). It should be noted that it is difficult to exclude experimental error in this comparison, because the single turnover kmax value does not require knowledge of the enzyme and DNA concentration whereas the calculated kcat value is dependent upon the concentration of enzyme and DNA substrate. However, we prefer an alternative explanation. It is expected that competitive binding sites on the DNA will decrease the binding of AlkA to the lesion site. The quantitative analysis of this level of competition is model specific (i.e., how many binding sites and what their affinities are relative to the lesion site), but qualitatively a 2-fold lower rate constant could be explained by AlkA spending ~50% of its time bound to non-damaged sites and 50% of its time bound to the damaged site. Consistent with this model, the affinity for undamaged DNA (Kd = 61 nM; Figure 5) is similar to the Kd value for excision of I•T from the 19u substrate (KM = 32 nM; Table 2). This indicates many alternative sites with much weaker affinity or a few alternative sites that are bound with similar affinity as damaged DNA. Crystal structures suggest that these competitive sites are simply the 5’ ends of the duplex (12, 13).
Although AlkA appears to dissociate from the Hx and abasic DNA products quickly, product inhibition is observed at points in the reaction where 10% conversion to product had occurred (See Supporting Information). We could find no evidence for inhibition of AlkA by 100 µM Hx and limited solubility precluded going to higher concentration (data not shown). In contrast, we observed potent inhibition by the abasic-site DNA product, with a Kd value of ~3 nM that is at least 10-fold higher affinity than for AlkA binding to an I•T lesion (Figure 5). This suggests that AlkA will bind tightly to any abasic sites that are present, even in the presence of broken DNA or weak-binding lesions such as I•T. Similar tight binding has been observed by many DNA glycosylases, and this is expected to protect abasic sites from deleterious reactions and to provide better coordination of the downstream reactions in the BER pathway.
Significance
In summary, we have provided biochemical evidence that AlkA binds tightly to DNA ends, supporting crystal structures of AlkA bound to DNA ends. Furthermore, we have identified a binding register that permits simple kinetic analysis under single turnover conditions. As most studies of AlkA have previously used symmetric substrates, it is likely that the activity of AlkA toward some substrates has been underestimated. It should be noted that E. coli has an alternative, faster pathway for repairing deaminated adenosine sites that is initiated by endonuclease V (Endo V) (34–36). Therefore, it is not clear if the activity of AlkA toward Hx lesions is under selective pressure in E. coli. Nevertheless, the rate enhancement for excision of Hx by AlkA is similar to that reported for both positively charged and neutral classes of substrates (10), suggesting that AlkA is not highly specialized for any of the known forms of DNA damage. The minimal kinetic mechanism presented here will serve as a framework for evaluating the activity of AlkA toward the exceptionally diverse group of damaged nucleotides that are substrates of AlkA.
Supplementary Material
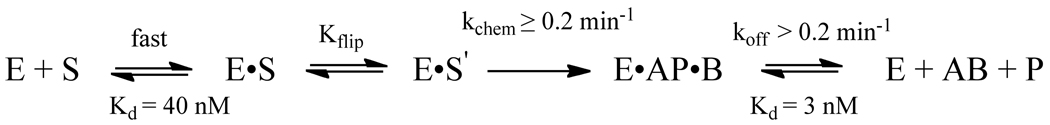
Figure 8.
Minimal kinetic mechanism for the recognition and excision of hypoxanthine by AlkA. The binding and flipping steps are assumed to be in rapid equilibrium, as has been seen for other glycosylases (32). The absence of a burst indicates that the release of Hx (B) and abasic DNA (AP) products are not rate limiting. However, AlkA shows potent inhibition by abasic sites. The rate of N-glycosidic bond cleavage must be at least as fast as the observed rate constant for the single turnover reaction, however it could be significantly faster if the flipping equilibrium is unfavorable (37).
ACKNOWLEDGMENT
We thank Michael Baldwin for purification of some of the oligonucleotides used in this study and members of the O’Brien lab for discussions and comments on the manuscript. B.Z. acknowledges the support of the Undergraduate Research Opportunities Program at the University of Michigan.
Abbreviations
- AlkA
Escherichia coli 3-methyladenine DNA glycosylase II
- BER
base excision repair
- BSA
bovine serum albumin
- εA
1,N6-ethenoadenine
- FAM
6-aminofluorescein
- Hx
hypoxanthine (the base moiety of inosine)
- I
2’-deoxyinosine
- NaHEPES
sodium N-(2-hydroxyethyl)piperazine-N’-2-ethanesulfonate
- NaMES
sodium 2-(N-morpholino)ethanesulfonate
- PAGE
polyacrylamide gel electrophoresis
Footnotes
This work was supported by a grant from the National Institutes of Health to P.O. (CA122254).
SUPPORTING INFORMATION AVAILABLE
Preincubation controls confirming the stability of AlkA, evidence for product inhibition, and additional data for the single turnover excision of εA and Hx. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Gilchrest BA, Bohr VA. Aging processes, DNA damage, and repair. FASEB J. 1997;11:322–330. doi: 10.1096/fasebj.11.5.9141498. [DOI] [PubMed] [Google Scholar]
- 3.Thomas L, Yang CH, Goldthwait DA. Two DNA glycosylases in Escherichia coli which release primarily 3-methyladenine. Biochemistry. 1982;21:1162–1169. doi: 10.1021/bi00535a009. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu. Rev. Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- 5.Saparbaev M, Kleibl K, Laval J. Escherichia coli, Saccharomyces cerevisiae, rat and human 3-methyladenine DNA glycosylases repair 1,N6-ethenoadenine when present in DNA. Nucleic Acids Res. 1995;23:3750–3755. doi: 10.1093/nar/23.18.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masaoka A, Terato H, Kobayashi M, Honsho A, Ohyama Y, Ide H. Enzymatic repair of 5-formyluracil. I. Excision of 5-formyluracil site-specifically incorporated into oligonucleotide substrates by alka protein (Escherichia coli 3-methyladenine DNA glycosylase II) J. Biol. Chem. 1999;274:25136–25143. doi: 10.1074/jbc.274.35.25136. [DOI] [PubMed] [Google Scholar]
- 7.Saparbaev M, Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc. Natl. Acad. Sci. U. S. A. 1994;91:5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjelland S, Bjoras M, Seeberg E. Excision of 3-methylguanine from alkylated DNA by 3-methyladenine DNA glycosylase I of Escherichia coli. Nucleic Acids Res. 1993;21:2045–2049. doi: 10.1093/nar/21.9.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terato H, Masaoka A, Asagoshi K, Honsho A, Ohyama Y, Suzuki T, Yamada M, Makino K, Yamamoto K, Ide H. Novel repair activities of AlkA (3-methyladenine DNA glycosylase II) and endonuclease VIII for xanthine and oxanine, guanine lesions induced by nitric oxide and nitrous acid. Nucleic Acids Res. 2002;30:4975–4984. doi: 10.1093/nar/gkf630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Brien PJ, Ellenberger T. The Escherichia coli 3-methyladenine DNA glycosylase AlkA has a remarkably versatile active site. J. Biol. Chem. 2004;279:26876–26884. doi: 10.1074/jbc.M403860200. [DOI] [PubMed] [Google Scholar]
- 11.Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowman BR, Lee S, Wang S, Verdine GL. Structure of the E. coli DNA glycosylase AlkA bound to the ends of duplex DNA: a system for the structure determination of lesion-containing DNA. Structure. 2008;16:1166–1174. doi: 10.1016/j.str.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S, Bowman BR, Ueno Y, Wang S, Verdine GL. Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine. J. Am. Chem. Soc. 2008;130:11570–11571. doi: 10.1021/ja8025328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowman BR, Lee S, Wang S, Verdine GL. Structure of Escherichia coli AlkA in complex with undamaged DNA. J. Biol. Chem. 2010;285:35783–35791. doi: 10.1074/jbc.M110.155663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Brien PJ, Ellenberger T. Human alkyladenine DNA glycosylase uses acid-base catalysis for selective excision of damaged purines. Biochemistry. 2003;42:12418–12429. doi: 10.1021/bi035177v. [DOI] [PubMed] [Google Scholar]
- 16.Hedglin M, O'Brien PJ. Human alkyladenine DNA glycosylase employs a processive search for DNA damage. Biochemistry. 2008;47:11434–11445. doi: 10.1021/bi801046y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldwin MR, O'Brien PJ. Human AP endonuclease I stimulates multiple-turnover base excision by alkyladenine DNA glycosylase. Biochemistry. 2009;48:6022–6033. doi: 10.1021/bi900517y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cleland WW. Substrate inhibition. Methods Enzymol. 1979;63:500–513. [PubMed] [Google Scholar]
- 19.McCullough AK, Scharer O, Verdine GL, Lloyd RS. Structural determinants for specific recognition by T4 endonuclease V. J. Biol. Chem. 1996;271:32147–32152. doi: 10.1074/jbc.271.50.32147. [DOI] [PubMed] [Google Scholar]
- 20.Scharer OD, Nash HM, Jiricny J, Laval J, Verdine GL. Specific binding of a designed pyrrolidine abasic site analog to multiple DNA glycosylases. J. Biol. Chem. 1998;273:8592–8597. doi: 10.1074/jbc.273.15.8592. [DOI] [PubMed] [Google Scholar]
- 21.Baldwin MR, O'Brien PJ. Nonspecific DNA binding and coordination of the first two steps of base excision repair. Biochemistry. 2010;49:7879–7891. doi: 10.1021/bi100889r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Stoerker J, Genuardi M, Yeung AT, Matsumoto Y, Bellacosa A. Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase. J. Biol. Chem. 2000;275:32422–32429. doi: 10.1074/jbc.M004535200. [DOI] [PubMed] [Google Scholar]
- 23.Waters TR, Gallinari P, Jiricny J, Swann PF. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
- 24.Porello SL, Leyes AE, David SS. Single-turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch-containing DNA substrates. Biochemistry. 1998;37:14756–14764. doi: 10.1021/bi981594+. [DOI] [PubMed] [Google Scholar]
- 25.Teo I, Sedgwick B, Kilpatrick MW, McCarthy TV, Lindahl T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell. 1986;45:315–324. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- 26.Barrett TE, Savva R, Barlow T, Brown T, Jiricny J, Pearl LH. Structure of a DNA base-excision product resembling a cisplatin inter-strand adduct. Nat. Struct. Biol. 1998;5:697–701. doi: 10.1038/1394. [DOI] [PubMed] [Google Scholar]
- 27.Wibley JE, Waters TR, Haushalter K, Verdine GL, Pearl LH. Structure and specificity of the vertebrate anti-mutator uracil-DNA glycosylase SMUG1. Mol. Cell. 2003;11:1647–1659. doi: 10.1016/s1097-2765(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 28.Grippon S, Zhao Q, Robinson T, Marshall JJ, O'Neill RJ, Manning H, Kennedy G, Dunsby C, Neil M, Halford SE, French PM, Baldwin GS. Differential modes of DNA binding by mismatch uracil DNA glycosylase from Escherichia coli: implications for abasic lesion processing and enzyme communication in the base excision repair pathway. Nucleic Acids Res. 2011;39:2593–2603. doi: 10.1093/nar/gkq913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maiti A, Morgan MT, Pozharski E, Drohat AC. Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8890–8895. doi: 10.1073/pnas.0711061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan MT, Maiti A, Fitzgerald ME, Drohat AC. Stoichiometry and affinity for thymine DNA glycosylase binding to specific and nonspecific DNA. Nucleic Acids Res. 2011;39:2319–2329. doi: 10.1093/nar/gkq1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fried MG, Kanugula S, Bromberg JL, Pegg AE. DNA binding mechanism of O6-alkylguanine-DNA alkyltransferase: stoichiometry and effects of DNA base composition and secondary structure on complex stability. Biochemistry. 1996;35:15295–15301. doi: 10.1021/bi960971k. [DOI] [PubMed] [Google Scholar]
- 32.Wolfe AE, O'Brien PJ. Kinetic mechanism for the flipping and excision of 1,N(6)-ethenoadenine by human alkyladenine DNA glycosylase. Biochemistry. 2009;48:11357–11369. doi: 10.1021/bi9015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maher RL, Vallur AC, Feller JA, Bloom LB. Slow base excision by human alkyladenine DNA glycosylase limits the rate of formation of AP sites and AP endonuclease 1 does not stimulate base excision. DNA Repair (Amst) 2007;6:71–81. doi: 10.1016/j.dnarep.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Sidorkina O, Saparbaev M, Laval J. Effects of nitrous acid treatment on the survival and mutagenesis of Escherichia coli cells lacking base excision repair (hypoxanthine-DNA glycosylase-ALK A protein) and/or nucleotide excision repair. Mutagenesis. 1997;12:23–28. doi: 10.1093/mutage/12.1.23. [DOI] [PubMed] [Google Scholar]
- 35.Weiss B. Removal of deoxyinosine from the Escherichia coli chromosome as studied by oligonucleotide transformation. DNA Repair (Amst) 2008;7:205–212. doi: 10.1016/j.dnarep.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao M, Hatahet Z, Melamede RJ, Kow YW. Purification and characterization of a novel deoxyinosine-specific enzyme, deoxyinosine 3' endonuclease, from Escherichia coli. J. Biol. Chem. 1994;269:16260–16268. [PubMed] [Google Scholar]
- 37.O'Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J. Biol. Chem. 2004;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.