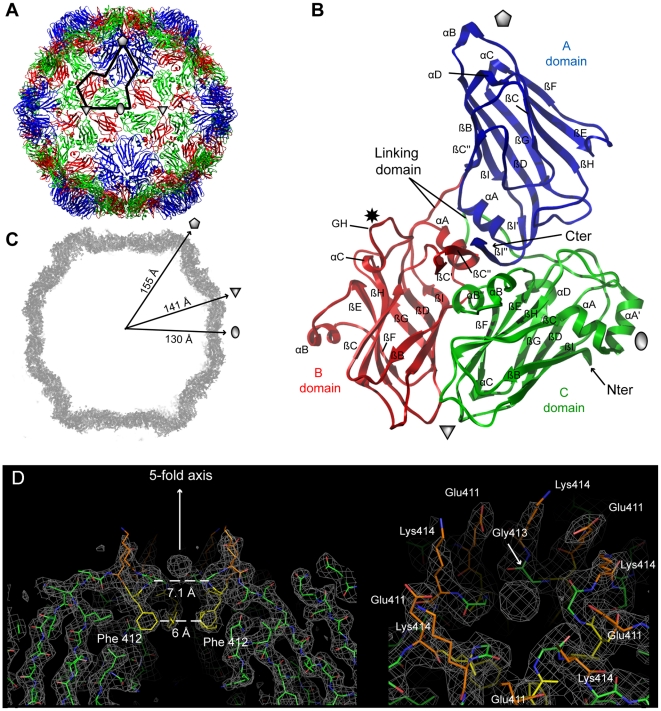
Figure 2. Crystal structures of GFLV-F13 and GFLV-TD.
(A) The structures of GFLV-F13 and of GFLV-TD are very similar as illustrated by the extremely low r.m.s.d. values (see Table S1). For this reason only the highest resolution model (GFLV-TD) is represented in this figure. The ribbon diagram of the virus capsid is viewed down an icosahedral 2-fold axis normal to the plane of the paper. Sixty copies of the CP are arranged in an icosahedral pseudo T = 3 symmetry. The black line delineates one CP position. The grey pentagon, triangle and oval symbolize the icosahedral 5-fold, 3-fold and 2-fold symmetry axes, respectively. (B) Each CP comprises three jellyroll β sandwiches termed C, B and A domains from the N- to the C-terminus and are depicted in green, red and blue, respectively. A star indicates the position of residue 297. (C) The central section of the 2Fo-Fc electron density map (2 σ contour level) of a GFLV-F13 particle is viewed down a viral 2-fold axis. The outer radial dimensions along the icosahedral symmetry axes are indicated. (D) A thin slice of 2Fo-Fc electron density map (contoured at 1σ) reveals a strong density peak (about 3 σ in the 2Fo-Fc and 17 σ in the Fo-Fc map) on the 5-fold axis of GFLV-TD. The arrow symbolizing the axis points towards the outer surface of the particle as well as neighbouring charged residues (Lys414 and Glu411) whereas Phe412 side chains are directed towards the viral cavity. The right panel shows a slightly shifted top view illustrating the organisation of the residues around the 5-fold axis. Five water molecules bridge Gly413 carbonyl groups and Asp411 side chains to the large central ion, possibly a phosphate coming from the crystallization medium.